Production, Passaging Stability, and Histological Analysis of Madin–Darby Canine Kidney Cells Cultured in a Low-Serum Medium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Study of MDCK Cell Culture and Production Process
2.2.1. Screening for Low-Serum Media
2.2.2. Validation of Scale-Up Culture
2.2.3. The Cell Production Process in a Low-Serum Medium
2.3. Passaging Stability of MDCK Cells in a Low Serum Medium
2.3.1. Stability of Continuously Passaged MDCK Cell Culture
2.3.2. Characterization of MDCK Cell Development across Generations
2.3.3. Virus Susceptibility Test
2.4. Transcriptomic and Proteomic Sequencing
2.4.1. RNA Extraction and RNA-Seq
2.4.2. Proteomic Analysis
3. Results
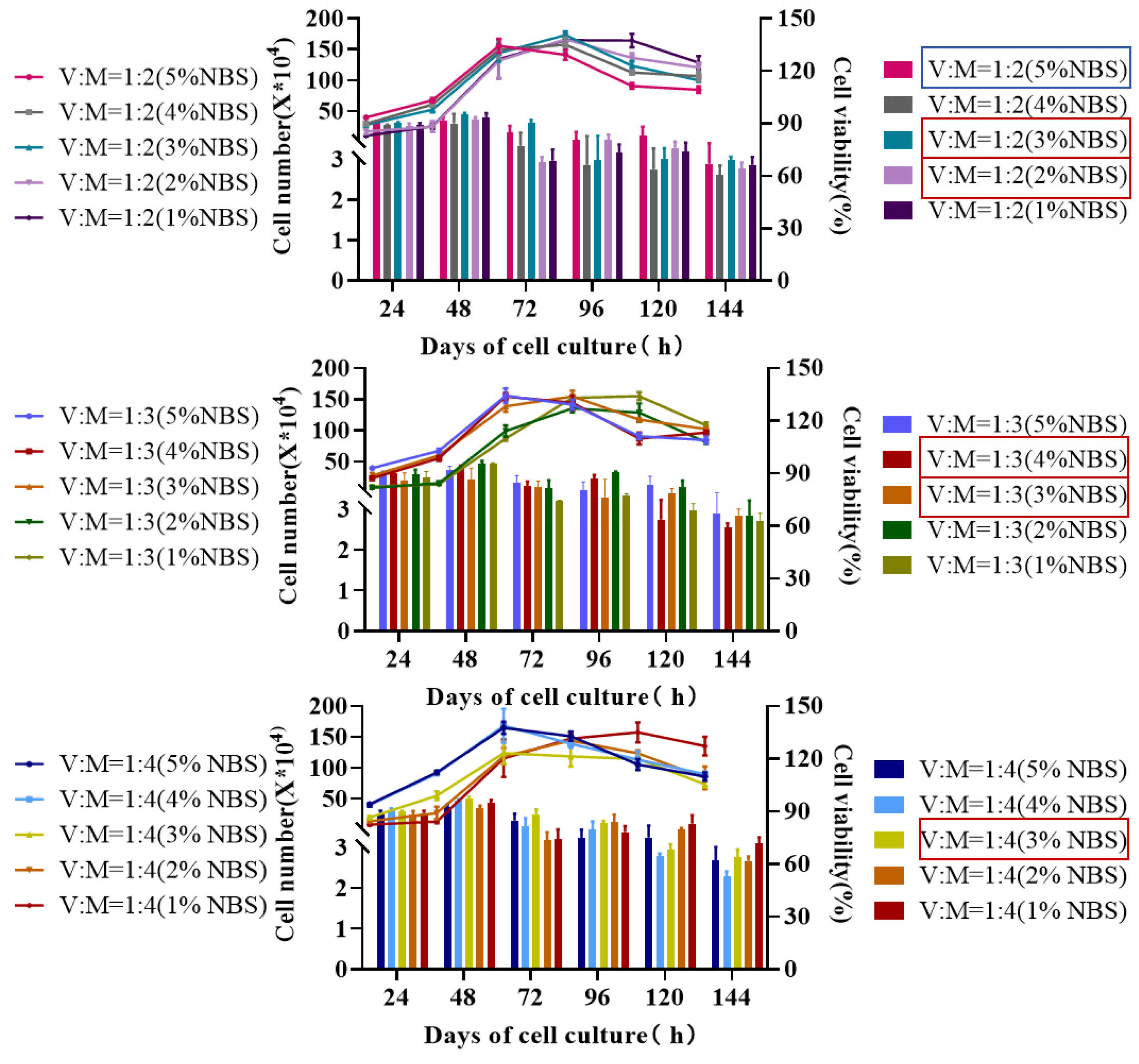
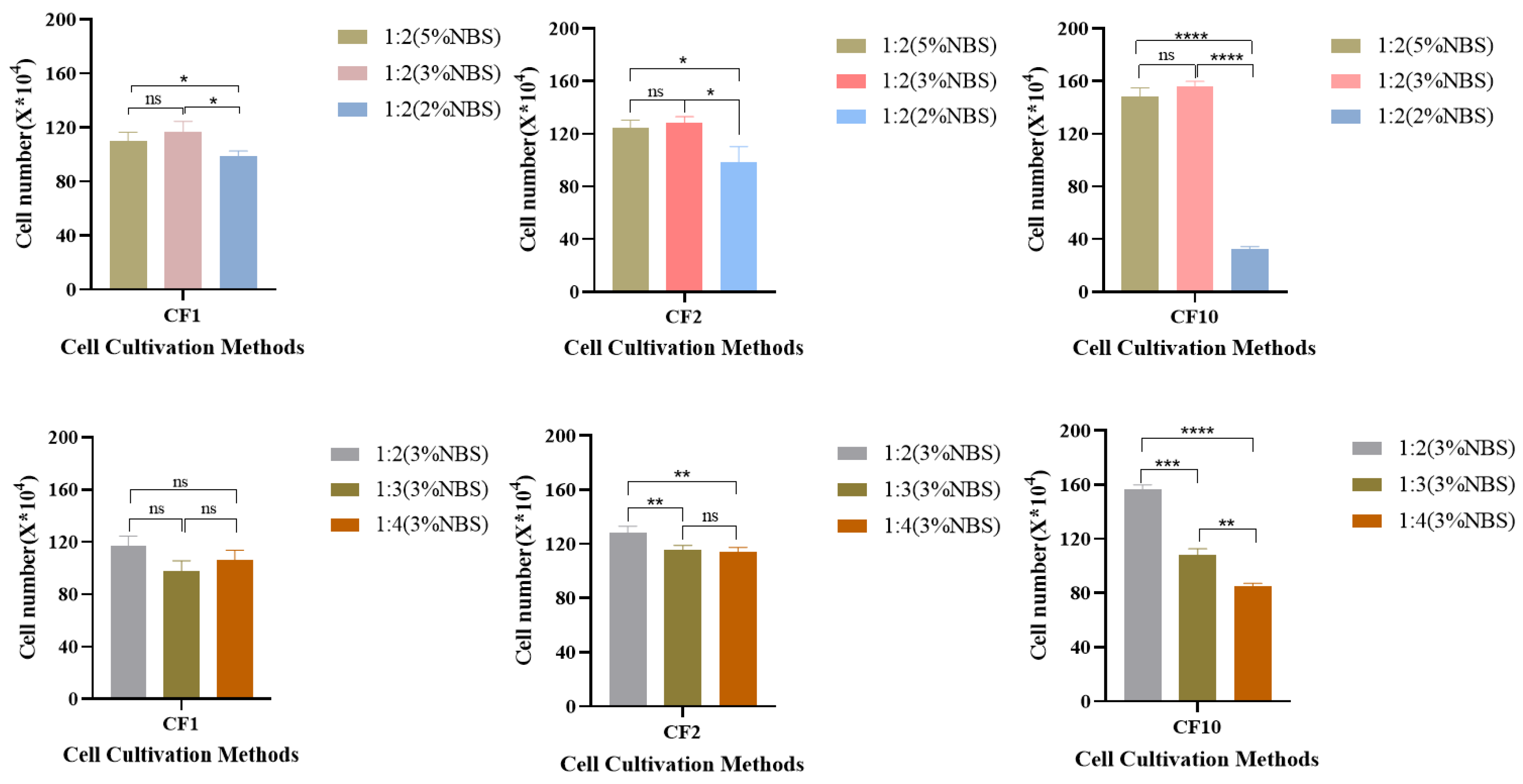
3.1. Study on the Culture Process of MDCK Cells in Low Serum Medium
3.2. Cell Culture in a Low-Serum Medium Bioreactor
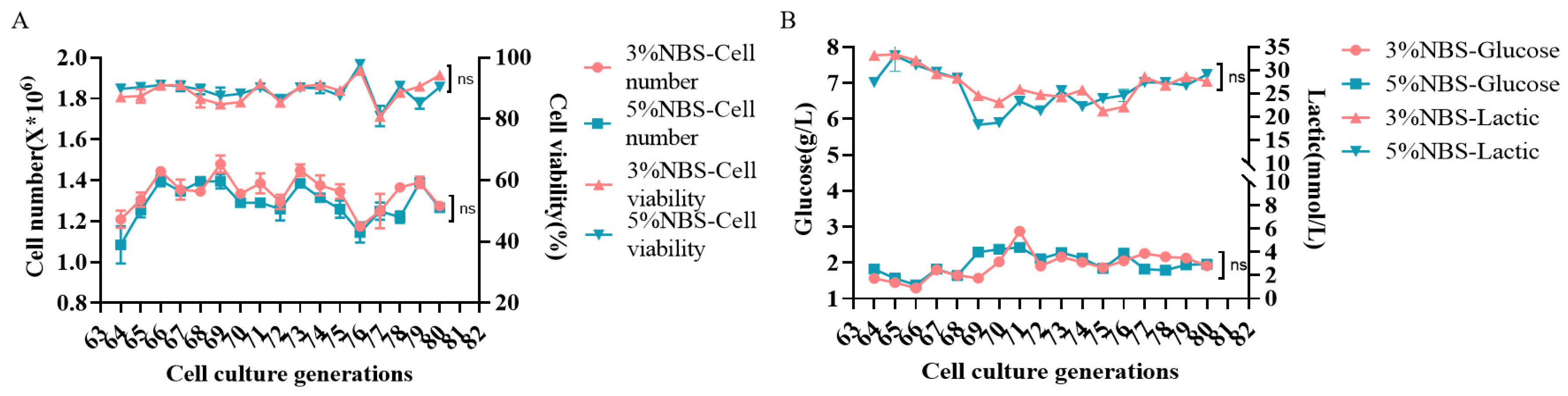
3.3. Stability Study of MDCK Cell Passaging
3.4. Transcriptomic and Proteomic Analysis of MDCK Cells Cultured in Medium with 3% and 5% Serum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pérez Rubio, A.; Eiros, J.M. Cell culture-derived flu vaccine: Present and future. Hum. Vaccines Immunother. 2018, 14, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Barr, I.G.; Donis, R.O.; Katz, J.M.; McCauley, J.W.; Odagiri, T.; Trusheim, H.; Tsai, T.F.; Wentworth, D.E. Cell culture-derived influenza vaccines in the severe 2017–2018 epidemic season: A step towards improved influenza vaccine effectiveness. NPJ Vaccines 2018, 3, 44. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-F.; Weng, T.-C.; Lai, C.-C.; Chen, P.-L.; Lee, M.-S.; Hu, A.Y.-C. A fast and efficient purification platform for cell-based influenza viruses by flow-through chromatography. Vaccine 2018, 36, 3146–3152. [Google Scholar] [CrossRef]
- Belser, J.A. Cell culture keeps pace with influenza virus. Lancet Respir. Med. 2018, 6, 805–806. [Google Scholar] [CrossRef]
- Rodrigues, A.F.; Fernandes, P.; Laske, T.; Castro, R.; Alves, P.M.; Genzel, Y.; Coroadinha, A.S. Cell Bank Origin of MDCK Parental Cells Shapes Adaptation to Serum-Free Suspension Culture and Canine Adenoviral Vector Production. Int. J. Mol. Sci. 2020, 21, 6111. [Google Scholar] [CrossRef]
- Haredy, A.M.; Takenaka, N.; Yamada, H.; Sakoda, Y.; Okamatsu, M.; Yamamoto, N.; Omasa, T.; Ohtake, H.; Mori, Y.; Kida, H.; et al. An MDCK cell culture-derived formalin-inactivated influenza virus whole-virion vaccine from an influenza virus library confers cross-protective immunity by intranasal administration in mice. Clin. Vaccine Immunol. 2013, 20, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Gaglani, M.; Kim, S.S.; Naleway, A.L.; Levine, M.Z.; Edwards, L.; Murthy, K.; Dunnigan, K.; Zunie, T.; Groom, H.; Ball, S.; et al. Effect of Repeat Vaccination on Immunogenicity of Quadrivalent Cell-Culture and Recombinant Influenza Vaccines Among Healthcare Personnel Aged 18–64 Years: A Randomized, Open-Label Trial. Clin. Infect. Dis. 2023, 76, e1168–e1176. [Google Scholar] [CrossRef] [PubMed]
- Nims, R.W.; Harbell, J.W. Best practices for the use and evaluation of animal serum as a component of cell culture medium. Vitr. Cell. Dev. Biol.—Anim. 2017, 53, 682–690. [Google Scholar] [CrossRef]
- Genzel, Y.; Olmer, R.M.; Schäfer, B.; Reichl, U. Wave microcarrier cultivation of MDCK cells for influenza virus pro-duction in serum containing and serum-free media. Vaccine 2006, 24, 6074–6087. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; Wen, L.N. LncRNA SNHG14 promotes proliferation of endometrial cancer through regulating mi-croRNA-655-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10410–10418. [Google Scholar] [PubMed]
- Peschel, B.; Frentzel, S.; Laske, T.; Genzel, Y.; Reichl, U. Comparison of influenza virus yields and apoptosis-induction in an adherent and a suspension MDCK cell line. Vaccine 2013, 31, 5693–5699. [Google Scholar] [CrossRef]
- Zhang, J.; Nian, X.; Liu, B.; Zhang, Z.; Zhao, W.; Han, X.; Ma, Y.; Jin, D.; Ma, H.; Zhang, Q.; et al. Development of MDCK-based quadrivalent split seasonal influenza virus vaccine with high safety and immunoprotection: A preclinical study. Antivir. Res. 2023, 216, 105639. [Google Scholar] [CrossRef]
- Divino, V.; Krishnarajah, G.; Pelton, S.I.; Mould-Quevedo, J.; Anupindi, V.R.; DeKoven, M.; Postma, M.J. A real-world study evaluating the relative vaccine effectiveness of a cell-based quadrivalent influenza vaccine compared to egg-based quadrivalent influenza vaccine in the US during the 2017-18 influenza season. Vaccine 2020, 38, 6334–6343. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.-Z.; Jiao, P.-R.; Qi, W.-B.; Fan, H.-Y.; Liao, M. Development and strategies of cell-culture technology for influenza vaccine. Appl. Microbiol. Biotechnol. 2011, 89, 893–902. [Google Scholar] [CrossRef]
- Aggarwal, K.; Jing, F.; Maranga, L.; Liu, J. Bioprocess optimization for cell culture based influenza vaccine production. Vaccine 2011, 29, 3320–3328. [Google Scholar] [CrossRef] [PubMed]
- Jochems, C.E.A.; van der Valk, J.B.; Stafleu, F.R.; Baumans, V. The use of fetal bovine serum: Ethical or scientific problem? Altern. Lab. Anim. 2002, 30, 219–227. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Guan, X.; Wang, S.; Liu, Q.; Chen, F.; Zheng, Y.; He, L.; Zhao, J. Establishment of Continuous In Vitro Culture of Babesia gibsoni by Using VP-SFM Medium with Low-Concentration Serum. Microbiol. Spectr. 2023, 11, e0025823. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, S.; Li, D.; Zhang, Y.; Luo, W.; Zhao, J.; He, L. Continuous In Vitro Culture of Babesia duncani in a Serum-Free Medium. Cells 2023, 12, 482. [Google Scholar] [CrossRef]
- Hassanzade, S.M.; Zavareh, A.; Shokrgozar, M.A.; Ramezani, A.; Fayaz, A. High vero cell density and rabies virus proliferation on fibracel disks versus cytodex-1 in spinner flask. Pak. J. Biol. Sci. 2011, 14, 441–448. [Google Scholar] [CrossRef]
- Chou, A.-H.; Liu, C.-C.; Chang, C.-P.; Guo, M.-S.; Hsieh, S.-Y.; Yang, W.-H.; Chao, H.-J.; Wu, C.-L.; Huang, J.-L.; Lee, M.-S.; et al. Pilot scale production of highly efficacious and stable enterovirus 71 vaccine candidates. PLoS ONE 2012, 7, e34834. [Google Scholar] [CrossRef]
- Taciak, B.; Białasek, M.; Braniewska, A.; Sas, Z.; Sawicka, P.; Kiraga, Ł.; Rygiel, T.; Król, M. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLoS ONE 2018, 13, e0198943. [Google Scholar] [CrossRef] [PubMed]
- Faqih, L.; Vallely, P.; Klapper, P. Genetic stability of SIV Gag/Tat gene inserted into Del-II in modified vaccinia virus ankara after serial passage of recombinant vector in pCEFs cells. J. Virol. Methods 2023, 312, 114651. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.J.; Saiz, J.C.; Laor, O.; Moore, D.M. Antigenic stability of foot-and-mouth disease virus variants on serial passage in cell culture. J. Virol. 1991, 65, 3949–3953. [Google Scholar] [CrossRef]
- Asai, D.; Kawano, T.; Murata, M.; Nakashima, H.; Toita, R.; Kang, J.-H. Effect of Fetal Bovine Serum Concentration on Lysophosphatidylcholine-mediated Proliferation and Apoptosis of Human Aortic Smooth Muscle Cells. J. Oleo Sci. 2020, 69, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Phan, T.; Hu, W.-S.; Liu, X.; Fan, L.; Tan, W.-S.; Zhao, L. Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses 2021, 13, 2200. [Google Scholar] [CrossRef]
- Liu, G.; Pei, M.; Wang, S.; Qiu, Z.; Li, X.; Ma, H.; Ma, Y.; Wang, J.; Qiao, Z.; Ma, Z.; et al. Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells. Vaccines 2023, 11, 1593. [Google Scholar] [CrossRef]
- Galassie, A.C.; Goll, J.B.; Samir, P.; Jensen, T.L.; Hoek, K.L.; Howard, L.M.; Allos, T.M.; Niu, X.; Gordy, L.E.; Creech, C.B.; et al. Proteomics show antigen presentation processes in human immune cells after AS03-H5N1 vaccination. Proteomics 2017, 17, 1600453. [Google Scholar] [CrossRef]
- Wimmers, F.; Donato, M.; Kuo, A.; Ashuach, T.; Gupta, S.; Li, C.; Dvorak, M.; Foecke, M.H.; Chang, S.E.; Hagan, T.; et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell 2021, 184, 3915–3935.e21. [Google Scholar] [CrossRef]
- Clark, J.M.; Gebb, C.; Hirtenstein, M.D. Serum supplements and serum-free media: Applicability for microcarrier culture of animal cells. Dev. Biol. Stand. 1981, 50, 81–91. [Google Scholar]
- Parker, A.; Shang, H.; Khurgel, M.; Katz, A. Low serum and serum-free culture of multipotential human adipose stem cells. Cytotherapy 2007, 9, 637–646. [Google Scholar] [CrossRef]
- Habibi, G.; Afshari, A.; Shahedi, A.; Hashemlou, M.; Es-Haghi, A.; Mehrabadi, M.H.F.; Fathi, S. Development of Low-Serum Culture Media for the in Vitro Cultivation of Theileria annulata S15 Cell Line. Iran. J. Parasitol. 2024, 19, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Dalzon, B.; Torres, A.; Devcic, J.; Fenel, D.; Sergent, J.-A.; Rabilloud, T. A Low-Serum Culture System for Prolonged in Vitro Toxicology Experiments on a Macrophage System. Front. Toxicol. 2021, 3, 780778. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Zhang, L.; Zhan, S.; Ge, W.; Tang, P. Investigation of proteome changes in osteoclastogenesis in low serum culture system using quantitative proteomics. Proteome Sci. 2016, 14, 8. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Li, M.; Jia, M.; Yang, L.; Wang, T.; Wang, Y.; Kang, L.; Li, M.; Kong, L. Transcriptome and proteomic analysis of mpox virus F3L-expressing cells. Front. Cell. Infect. Microbiol. 2024, 14, 1354410. [Google Scholar] [CrossRef]
- Takemon, Y.; Chick, J.M.; Gyuricza, I.G.; A Skelly, D.; Devuyst, O.; Gygi, S.P.; A Churchill, G.; Korstanje, R. Proteomic and transcriptomic profiling reveal different aspects of aging in the kidney. eLife 2021, 10, e62585. [Google Scholar] [CrossRef]
- Mikulski, R.; Domsic, J.F.; Ling, G.; Tu, C.; Robbins, A.H.; Silverman, D.N.; McKenna, R. Structure and catalysis by carbonic anhydrase II: Role of active-site tryptophan 5. Arch. Biochem. Biophys. 2011, 516, 97–102. [Google Scholar] [CrossRef]
- Lukasak, B.J.; Mitchener, M.M.; Kong, L.; Dul, B.E.; Lazarus, C.D.; Ramakrishnan, A.; Ni, J.; Shen, L.; Maze, I.; Muir, T.W. TGM2-mediated histone transglutamination is dictated by steric accessibility. Proc. Natl. Acad. Sci. USA 2022, 119, e2208672119. [Google Scholar] [CrossRef]
- Tian, B.; Yuan, Y.; Yang, Y.; Luo, Z.; Sui, B.; Zhou, M.; Fu, Z.F.; Zhao, L. Interferon-Inducible GTPase 1 Impedes the Dimerization of Rabies Virus Phosphoprotein and Restricts Viral Replication. J. Virol. 2020, 94, e01203-20. [Google Scholar] [CrossRef]
- Matusevich, O.; Egorov, V.; Gluzdikov, I.; Titov, M.; Zarubaev, V.; Shtro, A.; Slita, A.; Dukov, M.; Shurygina, A.-P.; Smirnova, T.; et al. Synthesis and antiviral activity of PB1 component of the influenza A RNA polymerase peptide fragments. Antivir. Res. 2015, 113, 4–10. [Google Scholar] [CrossRef]
- Ye, X.; Wang, J.; Qiao, Z.; Yang, D.; Wang, J.; Abudureyimu, A.; Yang, K.; Feng, Y.; Ma, Z.; Liu, Z. Quantitative proteomic analysis of MDCK cell adhesion. Mol. Omics 2021, 17, 121–129. [Google Scholar] [CrossRef]
- Yang, D.; Huang, L.; Wang, J.; Wu, H.; Liu, Z.; Abudureyimu, A.; Qiao, Z. Tumorigenesis mechanism and application strategy of the MDCK cell line: A systematic review. Biologicals 2023, 83, 101699. [Google Scholar] [CrossRef] [PubMed]



| VP:M199 (NBS) | 1:2 (3% NBS) | 1:2 (5% NBS) |
|---|---|---|
| Inoculation density | 80 × 104 cells/mL | 80 × 104 cells/mL |
| Culture time | 72 h | 72 h |
| Tank sugar concentration | 1.0 g/L | 1.0 g/L |
| Volume of medium | 20 L | 20 L |
| Number of cells after digestion | 278 × 104/mL × 4.5 L | 288 × 104/mL × 4.5 L |
| 102 × 104/mL × 4.5 L | 88 × 104/mL × 4.5 L | |
| 30 × 104/mL × 4.5 L | 32 × 104/mL × 4.5 L | |
| Total number of cells digested | 410 × 104/mL × 4.5 L | 408 × 104/mL × 4.5 L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, M.; Le, Y.; Gong, Z.; Dong, T.; Liu, B.; Su, M.; Li, X.; Peng, F.; Li, Q.; Nian, X.; et al. Production, Passaging Stability, and Histological Analysis of Madin–Darby Canine Kidney Cells Cultured in a Low-Serum Medium. Vaccines 2024, 12, 991. https://doi.org/10.3390/vaccines12090991
Cai M, Le Y, Gong Z, Dong T, Liu B, Su M, Li X, Peng F, Li Q, Nian X, et al. Production, Passaging Stability, and Histological Analysis of Madin–Darby Canine Kidney Cells Cultured in a Low-Serum Medium. Vaccines. 2024; 12(9):991. https://doi.org/10.3390/vaccines12090991
Chicago/Turabian StyleCai, Ming, Yang Le, Zheng Gong, Tianbao Dong, Bo Liu, Minne Su, Xuedan Li, Feixia Peng, Qingda Li, Xuanxuan Nian, and et al. 2024. "Production, Passaging Stability, and Histological Analysis of Madin–Darby Canine Kidney Cells Cultured in a Low-Serum Medium" Vaccines 12, no. 9: 991. https://doi.org/10.3390/vaccines12090991







