Response Type and Host Species may be Sufficient to Predict Dose-Response Curve Shape for Adenoviral Vector Vaccines
Abstract
:1. Introduction
- 1)
- assessing the prevalence of peaking/saturating dose-response curve shapes in published adenoviral vector vaccine studies.
- 2)
- assessing whether dose-response curve shape may be predicted by response type, host species, adenoviral species and route of administration (RoA).
- 3)
- assessing which of host species, adenoviral species and RoA are the most likely predictors of dose-response curve shape.
2. Materials and Methods
2.1. Data Collation and Preparation
2.2. Objective 1: Assessing the Prevalence of Peaking/Saturating Dose-Response Curve Shapes in Published Adenoviral Vector Vaccine Studies
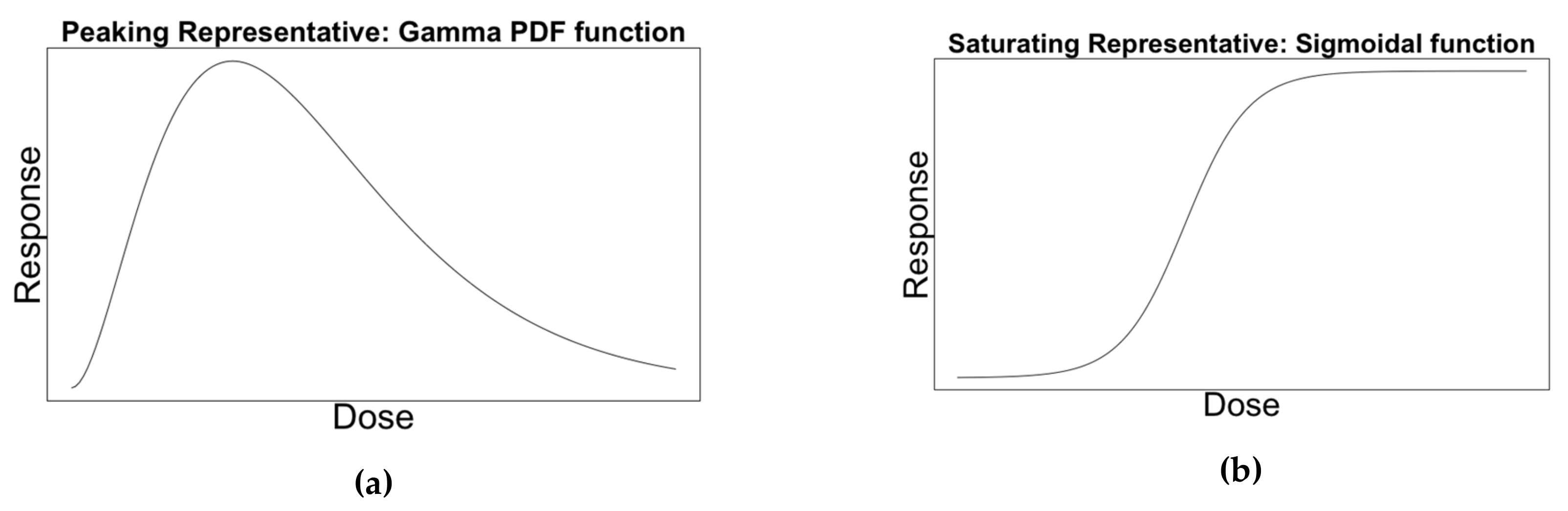
2.2.1. Representative Curves
2.2.2. Calibrating Curves to Data
- Provides no evidence, ΔAIC <2
- Positive evidence, 2 ≤ ΔAIC < 6
- Strong evidence, 6 ≤ ΔAIC < 10
- Very strong evidence 10 ≤ ΔAIC
2.2.3. Calculating Dose-Response Curve Shape Prevalence
2.2.4. Exploring Potential Bias of Independence Assumption
2.3. Objective 2: Assessing whether Dose-Response Curve Shape may be Predicted by Response Type, Host Species, Adenoviral Species, and Route of Administration (RoA)
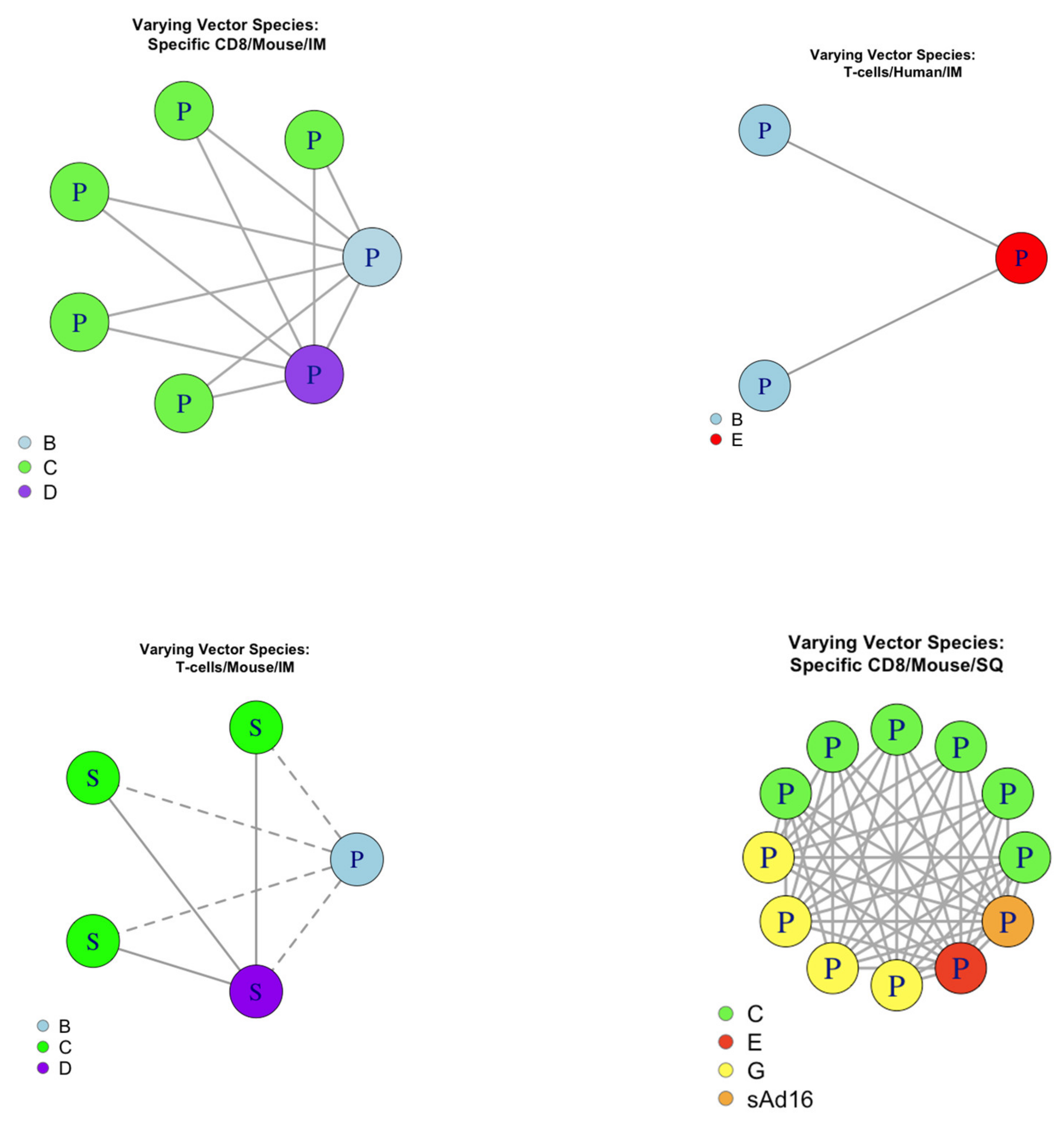
2.3.1. Grouping
2.3.2. Evaluating Consistency
2.4. Objective 3: Assessing which of Host Species, Adenoviral Species and RoA are the most likely Predictors of Dose-Response Curve Shape
2.4.1. Pairing
2.4.2. Evaluating Consistency
3. Results
3.1. Data
3.2. Objective 1: Assessing the Prevalence of Peaking/Saturating Dose-Response Curve Shapes in Published Adenoviral Vector Vaccine Studies
3.2.1. Overall Prevalence
3.2.2. Prevalence by Response Type
3.2.3. Prevalence in Human Data
3.2.4. Exploring Potential Bias of Independence Assumption
3.3. Objective 2: Assessing whether Dose-Response Curve Shape may be Predicted by Response Type, Host Species, Adenoviral Species, and Route of Administration (RoA).
Evaluating Consistency
3.4. Objective 3: Assessing which of Host Species, Adenoviral Species, and RoA are the most likely Predictors of Dose-Response Curve Shape
3.4.1. Evaluating Pairwise Consistency for Host
3.4.2. Evaluating pairwise consistency for adenoviral species
3.4.3. Evaluating pairwise consistency for RoA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Response Type | Vector Species | Host Species | Route of administration | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Papers | Antibodies | T cell | CD4+ | CD8+ | %CD4+ that is IFN+ | %CD8+ that is IFN+ | %CD4+ that is TNF+ | %CD8+ that is TNF+ | %CD4+ that is IL2+ | %CD8+ that is Il-2+ | %CD4+that is Il-17+ | Virus Neutralization Titre | B | C | D | E | G | None | Rabbit | Mouse | Human | Cattle | Monkey | Rat | SQ | IM |
| 140 [15] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 249 [16] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 305 [17] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 309 [18] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||
| 417 [19] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 441 [20] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||
| 461 [21] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 467 [22] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 555 [23] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
| 574 [24] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 578 [25] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 594 [26] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 633 [27] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 669 [28] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 686 [29] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 744 [30] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 924 [31] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 936 [32] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 1039 [33] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 1201 [34] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 1269 [35] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||
| 1343 [36] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 1474 [37] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 1492 [38] | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| 1539 [39] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 1801 [40] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 1877 [41] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 2030 [42] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 2505 [43] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 2531 [44] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 2841 [45] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 2916 [46] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 2919 [47] | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 2980 [48] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| 3018 [49] | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| Sum | 18 | 12 | 4 | 12 | 2 | 3 | 2 | 1 | 2 | 1 | 1 | 11 | 9 | 18 | 4 | 9 | 1 | 1 | 1 | 21 | 9 | 1 | 2 | 2 | 5 | |
References
- Andre, F.; Booy, R.; Bock, H.; Clemens, J.; Datta, S.; John, T.; Lee, B.; Lolekha, S.; Peltola, H.; Ruff, T.; et al. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull. World Health Organ. 2008, 86, 140–146. [Google Scholar] [CrossRef]
- Fougeroux, C.; Holst, P.J. Future Prospects for the Development of Cost-Effective Adenovirus Vaccines. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhou, D. Adenoviral vector-based strategies against infectious disease and cancer. Hum. Vaccines Immunother. 2016, 12, 2064–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, S.J.; Knight, G.M.; Kirschner, D.E.; White, R.G.; Evans, T.G. Dose finding for new vaccines: The role for immunostimulation/immunodynamic modelling. J. Theor. Biol. 2018. ArXiv181104024 Q-Bio. [Google Scholar] [CrossRef] [PubMed]
- Beals, C.R.; Railkar, R.A.; Schaeffer, A.K.; Levin, Y.; Kochba, E.; Meyer, B.K.; Evans, R.K.; Sheldon, E.A.; Lasseter, K.; Lang, N.; et al. Immune response and reactogenicity of intradermal administration versus subcutaneous administration of varicella-zoster virus vaccine: An exploratory, randomised, partly blinded trial. Lancet Infect. Dis. 2016, 16, 915–922. [Google Scholar] [CrossRef]
- Rhodes, S.J.; Guedj, J.; Fletcher, H.A.; Lindenstrøm, T.; Scriba, T.J.; Evans, T.G.; Knight, G.M.; White, R.G. Using vaccine Immunostimulation/Immunodynamic modelling methods to inform vaccine dose decision-making. NPJ Vaccines 2018, 3, 36. [Google Scholar] [CrossRef]
- Handel, A.; Li, Y.; McKay, B.; Pawelek, K.A.; Zarnitsyna, V.; Antia, R. Exploring the impact of inoculum dose on host immunity and morbidity to inform model-based vaccine design. PLOS Comput. Biol. 2018, 14, e1006505. [Google Scholar] [CrossRef] [Green Version]
- Rhodes, S.J.; Zelmer, A.; Knight, G.M.; Prabowo, S.A.; Stockdale, L.; Evans, T.G.; Lindenstrøm, T.; White, R.G.; Fletcher, H. The TB vaccine H56+IC31 dose-response curve is peaked not saturating: Data generation for new mathematical modelling methods to inform vaccine dose decisions. Vaccine 2016, 34, 6285–6291. [Google Scholar] [CrossRef] [Green Version]
- Afrough, S.; Rhodes, S.; Evans, T.; White, R.; Benest, J. Immunologic Dose-Response to Adenovirus-Vectored Vaccines in Animals and Humans: A Systematic Review of Dose-Response Studies of Replication Incompetent Adenoviral Vaccine Vectors when Given via an Intramuscular or Subcutaneous Route. Vaccines 2020, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Dicks, M.D.J.; Spencer, A.J.; Edwards, N.J.; Wadell, G.; Bojang, K.; Gilbert, S.C.; Hill, A.V.S.; Cottingham, M.G. A Novel Chimpanzee Adenovirus Vector with Low Human Seroprevalence: Improved Systems for Vector Derivation and Comparative Immunogenicity. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Grothendieck, G. nls2: Non-linear regression with brute force; R package version 0.2. 2013. Available online: https://CRAN.R-project.org/package=nls2 (accessed on 5 January 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Ciaburro, G. Regression Analysis with R: Design and Develop Statistical Nodes to Identify Unique Relationships Within Data at Scale; Packt Publishing Ltd.: Birmingham, UK, 2018; ISBN 978-1-78862-270-7. [Google Scholar]
- Raftery, A.E. Bayesian Model Selection in Social Research. Sociol. Methodol. 1995, 25, 111–163. [Google Scholar] [CrossRef]
- Tapia, M.D.; Sow, S.O.; Lyke, K.E.; Haidara, F.C.; Diallo, F.; Doumbia, M.; Traore, A.; Coulibaly, F.; Kodio, M.; Onwuchekwa, U.; et al. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: A phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2016, 16, 31–42. [Google Scholar] [PubMed] [Green Version]
- Ewer, K.; Rampling, T.; Venkatraman, N.; Bowyer, G.; Wright, D.; Lambe, T.; Imoukhuede, E.B.; Payne, R.; Fehling, S.K.; Strecker, T.; et al. A Monovalent Chimpanzee Adenovirus Ebola Vaccine Boosted with MVA. N. Engl. J. Med. 2016, 374, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Ondondo, B.; Abdul-Jawad, S.; Bridgeman, A.; Hanke, T. Characterization of T-Cell Responses to Conserved Regions of the HIV-1 Proteome in BALB/c Mice. Clin. Vaccine Immunol. CVI 2014, 21, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Kagina, B.M.N.; Tameris, M.D.; Geldenhuys, H.; Hatherill, M.; Abel, B.; Hussey, G.D.; Scriba, T.J.; Mahomed, H.; Sadoff, J.C.; Hanekom, W.A.; et al. The novel tuberculosis vaccine, AERAS-402, is safe in healthy infants previously vaccinated with BCG, and induces dose-dependent CD4 and CD8T cell responses. Vaccine 2014, 32, 5908–5917. [Google Scholar] [CrossRef]
- Antrobus, R.D.; Coughlan, L.; Berthoud, T.K.; Dicks, M.D.; Hill, A.V.; Lambe, T.; Gilbert, S.C. Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved Influenza A antigens. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 668–674. [Google Scholar] [CrossRef] [Green Version]
- Ouédraogo, A.; Tiono, A.B.; Kargougou, D.; Yaro, J.B.; Ouédraogo, E.; Kaboré, Y.; Kangoye, D.; Bougouma, E.C.; Gansane, A.; Henri, N.; et al. A phase 1b randomized, controlled, double-blinded dosage-escalation trial to evaluate the safety, reactogenicity and immunogenicity of an adenovirus type 35 based circumsporozoite malaria vaccine in Burkinabe healthy adults 18 to 45 years of age. PLoS ONE 2013, 8, e78679. [Google Scholar] [CrossRef]
- Vemula, S.V.; Amen, O.; Katz, J.M.; Donis, R.; Sambhara, S.; Mittal, S.K. Beta-defensin 2 enhances immunogenicity and protection of an adenovirus-based H5N1 influenza vaccine at an early time. Virus Res. 2013, 178, 398–403. [Google Scholar] [CrossRef] [Green Version]
- Creech, C.B.; Dekker, C.L.; Ho, D.; Phillips, S.; Mackey, S.; Murray-Krezan, C.; Grazia Pau, M.; Hendriks, J.; Brown, V.; Dally, L.G.; et al. Randomized, placebo-controlled trial to assess the safety and immunogenicity of an adenovirus type 35-based circumsporozoite malaria vaccine in healthy adults. Hum. Vaccines Immunother. 2013, 9, 2548–2557. [Google Scholar] [CrossRef] [Green Version]
- Quinn, K.M.; Da Costa, A.; Yamamoto, A.; Berry, D.; Lindsay, R.W.B.; Darrah, P.A.; Wang, L.; Cheng, C.; Kong, W.-P.; Gall, J.G.D.; et al. Comparative analysis of the magnitude, quality, phenotype, and protective capacity of simian immunodeficiency virus gag-specific CD8+ T cells following human-, simian-, and chimpanzee-derived recombinant adenoviral vector immunization. J. Immunol. Baltim. Md 1950 2013, 190, 2720–2735. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Wang, H.; Wang, F.; Yu, L. Recombinant adenovirus expressing type Asia1 foot-and-mouth disease virus capsid proteins induces protective immunity against homologous virus challenge in mice. Res. Vet. Sci. 2013, 94, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Zahn, R.; Gillisen, G.; Roos, A.; Koning, M.; van der Helm, E.; Spek, D.; Weijtens, M.; Grazia Pau, M.; Radošević, K.; Weverling, G.J.; et al. Ad35 and ad26 vaccine vectors induce potent and cross-reactive antibody and T-cell responses to multiple filovirus species. PLoS ONE 2012, 7, e44115. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; Walsh, S.R.; Seaman, M.S.; Tucker, R.P.; Krause, K.H.; Patel, A.; Johnson, J.A.; Kleinjan, J.; Yanosick, K.E.; Perry, J.; et al. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J. Infect. Dis. 2013, 207, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Keefer, M.C.; Gilmour, J.; Hayes, P.; Gill, D.; Kopycinski, J.; Cheeseman, H.; Cashin-Cox, M.; Naarding, M.; Clark, L.; Fernandez, N.; et al. A phase I double blind, placebo-controlled, randomized study of a multigenic HIV-1 adenovirus subtype 35 vector vaccine in healthy uninfected adults. PLoS ONE 2012, 7, e41936. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Singh, N.; Vemula, S.V.; Couëtil, L.; Katz, J.M.; Donis, R.; Sambhara, S.; Mittal, S.K. Impact of preexisting adenovirus vector immunity on immunogenicity and protection conferred with an adenovirus-based H5N1 influenza vaccine. PLoS ONE 2012, 7, e33428. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, G.A.; Duncan, C.J.A.; Ewer, K.J.; Collins, K.A.; Elias, S.C.; Halstead, F.D.; Goodman, A.L.; Edwards, N.J.; Reyes-Sandoval, A.; Bird, P.; et al. Clinical assessment of a recombinant simian adenovirus ChAd63: A potent new vaccine vector. J. Infect. Dis. 2012, 205, 772–781. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.-Y.; Tian, D.-Y.; Han, Q.-Y.; Zhang, X.; Li, N.; Qiu, H.-J. A novel alphavirus replicon-vectored vaccine delivered by adenovirus induces sterile immunity against classical swine fever. Vaccine 2011, 29, 8364–8372. [Google Scholar] [CrossRef]
- Rhee, E.G.; Kelley, R.P.; Agarwal, I.; Lynch, D.M.; Porte, A.L.; Simmons, N.L.; Clark, S.L.; Barouch, D.H. TLR4 Ligands Augment Antigen-Specific CD8+ T Lymphocyte Responses Elicited by a Viral Vaccine Vector. J. Virol. 2010, 84, 10413–10419. [Google Scholar] [CrossRef] [Green Version]
- Steitz, J.; Barlow, P.G.; Hossain, J.; Kim, E.; Okada, K.; Kenniston, T.; Rea, S.; Donis, R.O.; Gambotto, A. A Candidate H1N1 Pandemic Influenza Vaccine Elicits Protective Immunity in Mice. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [Green Version]
- Richardson, J.S.; Yao, M.K.; Tran, K.N.; Croyle, M.A.; Strong, J.E.; Feldmann, H.; Kobinger, G.P. Enhanced protection against Ebola virus mediated by an improved adenovirus-based vaccine. PLoS ONE 2009, 4, e5308. [Google Scholar] [CrossRef]
- Rodríguez, A.; Goudsmit, J.; Companjen, A.; Mintardjo, R.; Gillissen, G.; Tax, D.; Sijtsma, J.; Weverling, G.J.; Holterman, L.; Lanar, D.E.; et al. Impact of Recombinant Adenovirus Serotype 35 Priming versus Boosting of a Plasmodium falciparum Protein: Characterization of T- and B-Cell Responses to Liver-Stage Antigen 1. Infect. Immun. 2008, 76, 1709–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ophorst, O.J.A.E.; Radosević, K.; Klap, J.M.; Sijtsma, J.; Gillissen, G.; Mintardjo, R.; van Ooij, M.J.M.; Holterman, L.; Companjen, A.; Goudsmit, J.; et al. Increased immunogenicity of recombinant Ad35-based malaria vaccine through formulation with aluminium phosphate adjuvant. Vaccine 2007, 25, 6501–6510. [Google Scholar] [CrossRef] [PubMed]
- Widjojoatmodjo, M.N.; Bogaert, L.; Meek, B.; Zahn, R.; Vellinga, J.; Custers, J.; Serroyen, J.; Radošević, K.; Schuitemaker, H. Recombinant low-seroprevalent adenoviral vectors Ad26 and Ad35 expressing the respiratory syncytial virus (RSV) fusion protein induce protective immunity against RSV infection in cotton rats. Vaccine 2015, 33, 5406–5414. [Google Scholar] [CrossRef] [Green Version]
- Capone, S.; Meola, A.; Ercole, B.B.; Vitelli, A.; Pezzanera, M.; Ruggeri, L.; Davies, M.E.; Tafi, R.; Santini, C.; Luzzago, A.; et al. A novel adenovirus type 6 (Ad6)-based hepatitis C virus vector that overcomes preexisting anti-ad5 immunity and induces potent and broad cellular immune responses in rhesus macaques. J. Virol. 2006, 80, 1688–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ophorst, O.J.A.E.; Radošević, K.; Havenga, M.J.E.; Pau, M.G.; Holterman, L.; Berkhout, B.; Goudsmit, J.; Tsuji, M. Immunogenicity and Protection of a Recombinant Human Adenovirus Serotype 35-Based Malaria Vaccine against Plasmodium yoelii in Mice. Infect. Immun. 2006, 74, 313–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, M.; Boyer, J.L.; Hackett, N.R.; Wilson, J.M.; Crystal, R.G. Induction of Protective Immunity to Anthrax Lethal Toxin with a Nonhuman Primate Adenovirus-Based Vaccine in the Presence of Preexisting Anti-Human Adenovirus Immunity. Infect. Immun. 2005, 73, 6885–6891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, A.R.; Fitzgerald, J.C.; Giles-Davis, W.; Gao, G.P.; Wilson, J.M.; Ertl, H.C.J. Induction of CD8+ T cells to an HIV-1 antigen through a prime boost regimen with heterologous E1-deleted adenoviral vaccine carriers. J. Immunol. Baltim. Md 1950 2003, 171, 6774–6779. [Google Scholar] [CrossRef]
- Casimiro, D.R.; Chen, L.; Fu, T.-M.; Evans, R.K.; Caulfield, M.J.; Davies, M.-E.; Tang, A.; Chen, M.; Huang, L.; Harris, V.; et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J. Virol. 2003, 77, 6305–6313. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Z.; Gao, G.; Reyes-Sandoval, A.; Cohen, C.J.; Li, Y.; Bergelson, J.M.; Wilson, J.M.; Ertl, H.C.J. Novel, Chimpanzee Serotype 68-Based Adenoviral Vaccine Carrier for Induction of Antibodies to a Transgene Product. J. Virol. 2002, 76, 2667–2675. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Z.Q.; Yang, Y.; Wilson, J.M.; Ertl, H.C. A replication-defective human adenovirus recombinant serves as a highly efficacious vaccine carrier. Virology 1996, 219, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Eloit, M.; Adam, M. Isogenic adenoviruses type 5 expressing or not expressing the E1A gene: Efficiency as virus vectors in the vaccination of permissive and non-permissive species. J. Gen. Virol. 1995, 76, 1583–1589. [Google Scholar] [CrossRef] [PubMed]
- Neilan, J.G.; Schutta, C.; Barrera, J.; Pisano, M.; Zsak, L.; Hartwig, E.; Rasmussen, M.V.; Kamicker, B.J.; Ettyreddy, D.; Brough, D.E.; et al. Efficacy of an adenovirus-vectored foot-and-mouth disease virus serotype A subunit vaccine in cattle using a direct contact transmission model. BMC Vet. Res. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.; Freed, D.C.; Li, F.; Meschino, S.; Prokop, M.; Bett, A.; Casimiro, D.; Wang, D.; Fu, T.-M. Functionally inactivated dominant viral antigens of human cytomegalovirus delivered in replication incompetent adenovirus type 6 vectors as vaccine candidates. Hum. Vaccines Immunother. 2017, 13, 2763–2771. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, J.A.; McCaffery, J.N.; Kashentseva, E.; Singh, B.; Dmitriev, I.; Curiel, D.T.; Moreno, A. A prime-boost immunization regimen based on a Simian Adenovirus 36 vectored multi-stage malaria vaccine induces protective immunity in mice. Vaccine 2017, 35, 3239–3248. [Google Scholar] [CrossRef]
- Nazerai, L.; Bassi, M.R.; Uddback, I.E.M.; Holst, P.J.; Christensen, J.P.; Thomsen, A.R. Early life vaccination: Generation of adult-quality memory CD8+ T cells in infant mice using non-replicating adenoviral vectors. Sci. Rep. 2016, 6, 38666. [Google Scholar] [CrossRef]
- Penaloza-MacMaster, P.; Alayo, Q.A.; Ra, J.; Provine, N.M.; Larocca, R.; Lee, B.; Barouch, D.H. Inhibitory receptor expression on memory CD8 T cells following Ad vector immunization. Vaccine 2016, 34, 4955–4963. [Google Scholar] [CrossRef] [Green Version]




| Total Number of All Datasets by Level of Evidence and Response | ||||||||
|---|---|---|---|---|---|---|---|---|
| Response Type | Very Strong Peaking 10≤ ΔAIC | Strong Peaking 6≤ ΔAIC <10 | Positive Peaking 2≤ ΔAIC <6 | No Evidence ΔAIC<2 | Positive Saturating 2≤ ΔAIC <6 | Strong Saturating 6≤ ΔAIC <10 | Very Strong Saturating 10≤ ΔAIC | Total Number of Datasets for Response |
| Antibody | 3 | 0 | 2 | 46 | 1 | 1 | 1 | 54 |
| T-Cell | 1 | 2 | 0 | 27 | 3 | 1 | 0 | 34 |
| CD4+ | 1 | 1 | 0 | 4 | 0 | 0 | 0 | 6 |
| CD8+ | 11 | 3 | 7 | 42 | 0 | 0 | 0 | 63 |
| CD4 IFNγ+ | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| CD8 IFNγ+ | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 3 |
| CD4+ TNFα+ | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| CD8+ TNFα+ | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| CD4+ IL-2+ | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| CD8+ IL-2+ | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| CD4+ IL-17+ | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Virus Neutralization Titre | 2 | 0 | 1 | 17 | 1 | 1 | 0 | 22 |
| Total that provided evidence (%) | 42 (22.0%) | 140 (73.3%) | 8 (4.7%) | 191 | ||||
| Total Number of Human Datasets by Associated Level of Evidence | |||||||
|---|---|---|---|---|---|---|---|
| Very Strong Peaking 10≤ΔAIC | Strong Peaking 6≤ΔAIC<10 | Positive Peaking 2≤ΔAIC<6 | No Evidence ΔAIC<2 | Positive Saturating 2≤ΔAIC<6 | Strong Saturating 6≤ΔAIC<10 | Very Strong Saturating 10≤ΔAIC | |
| All responses (%) | 6 (16.2%) | 4 (10.8%) | 6 (16.2%) | 21 (56.8%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Total that provided evidence (%) | 16 (43.2%) | 21 (56.8%) | 0 (0%) | ||||
| Group (Response Type /Host/Adenoviral Species/Route of Administration) | Number of Datasets Providing Evidence Towards a Peaking Shape | Number of Datasets Providing Evidence Towards a Saturating Shape | Consistency |
|---|---|---|---|
| Antibodies/Human/B/IM | 2 | 0 | Consistent |
| Antibodies/Mouse/C/IM | 2 | 0 | Consistent |
| Antibodies/Monkey/C/IM | 0 | 3 | Consistent |
| T Cell/Mouse/C/IM | 0 | 3 | Consistent |
| CD4+/Human/B/IM | 2 | 0 | Consistent |
| CD8+/Human/B/IM | 2 | 0 | Consistent |
| CD8+/Mouse/C/IM | 5 | 0 | Consistent |
| CD8+/Mouse/C/SQ | 6 | 0 | Consistent |
| CD8+/Mouse/G/SQ | 4 | 0 | Consistent |
| CD4+ IFNγ+/Human/B/IM | 2 | 0 | Consistent |
| Virus Neutralization Titre/Mouse/C/IM | 2 | 0 | Consistent |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benest, J.; Rhodes, S.; Afrough, S.; Evans, T.; White, R. Response Type and Host Species may be Sufficient to Predict Dose-Response Curve Shape for Adenoviral Vector Vaccines. Vaccines 2020, 8, 155. https://doi.org/10.3390/vaccines8020155
Benest J, Rhodes S, Afrough S, Evans T, White R. Response Type and Host Species may be Sufficient to Predict Dose-Response Curve Shape for Adenoviral Vector Vaccines. Vaccines. 2020; 8(2):155. https://doi.org/10.3390/vaccines8020155
Chicago/Turabian StyleBenest, John, Sophie Rhodes, Sara Afrough, Thomas Evans, and Richard White. 2020. "Response Type and Host Species may be Sufficient to Predict Dose-Response Curve Shape for Adenoviral Vector Vaccines" Vaccines 8, no. 2: 155. https://doi.org/10.3390/vaccines8020155






