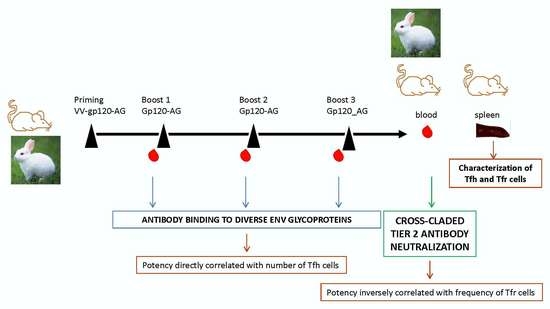
A Prime-Boost Immunization Strategy with Vaccinia Virus Expressing Novel gp120 Envelope Glycoprotein from a CRF02_AG Isolate Elicits Cross-Clade Tier 2 HIV-1 Neutralizing Antibodies
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells, Plasmids, Viruses, and Antibodies
2.3. Cloning of Envelope Genes from Primary Isolates
2.4. Production of Recombinant Vaccinia Viruses
2.5. Expression and Purification of Truncated gp120 Env Glycoproteins
2.6. Antigenic Characterization of Envelope Glycoproteins
2.7. Production, Expression, and Analysis of C2V3C3 Polypeptides
2.8. BALB/c Mice Immunizations
2.9. Envelope-specific Antibody Binding Reactivity in Sera of Immunized Animals
2.10. Neutralization Assays
2.11. Statistical Analysis
2.12. Accession Number(s)
3. Results
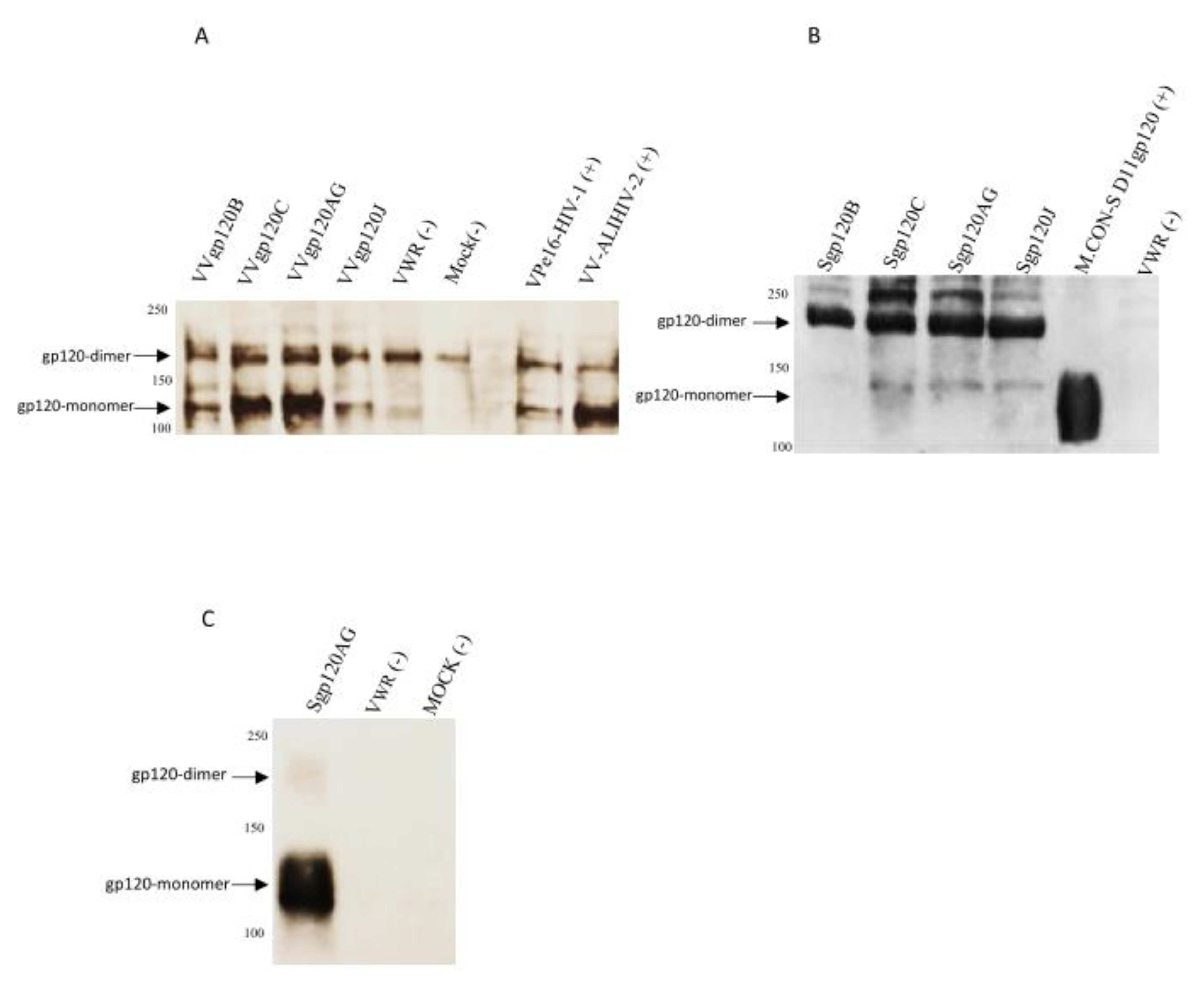
3.1. Generation of Recombinant Vaccinia Viruses Expressing gp120t from Different HIV-1 Clades
3.2. New Envelope Glycoproteins Bind to V3-specific and CD4bs-specific Neutralizing Monoclonal Antibodies
3.3. Antigenicity of the C2V3C3 Polypeptide
3.4. Envelope Glycoproteins Elicit Cross-reactive gp120-specific Binding Antibodies in Immunized Animals
3.5. Production of Heterologous Tier 2 HIV-1 Neutralizing Antibodies in Immunized Animals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stephenson, K.E.; D’Couto, H.T.; Barouch, D.H. New concepts in HIV-1 vaccine development. Curr. Opin. Immunol. 2016, 41, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Verkoczy, L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science 2014, 344, 588–589. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; van Gils, M.J.; Derking, R.; Sok, D.; Ketas, T.J.; Burger, J.A.; Ozorowski, G.; Cupo, A.; Simonich, C.; Goo, L.; et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015, 349, aac4223. [Google Scholar] [CrossRef] [PubMed]
- Sadanand, S.; Suscovich, T.J.; Alter, G. Broadly Neutralizing Antibodies Against HIV: New Insights to Inform Vaccine Design. Annu. Rev. Med. 2016, 67, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.M.; Geller, R.; Garijo, R.; Lopez-Aldeguer, J.; Sanjuan, R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015, 13, e1002251. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Mouquet, H.; Dosenovic, P.; Scheid, J.F.; Scharf, L.; Nussenzweig, M.C. Antibodies in HIV-1 vaccine development and therapy. Science 2013, 341, 1199–1204. [Google Scholar] [CrossRef]
- Peeters, M.; Jung, M.; Ayouba, A. The origin and molecular epidemiology of HIV. Expert. Rev. Anti. Infect. Ther. 2013, 11, 885–896. [Google Scholar] [CrossRef]
- Mascola, J.R.; Haynes, B.F. HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol. Rev. 2013, 254, 225–244. [Google Scholar] [CrossRef]
- van Gils, M.J.; Sanders, R.W. Broadly neutralizing antibodies against HIV-1: Templates for a vaccine. Virology 2013, 435, 46–56. [Google Scholar] [CrossRef]
- Montefiori, D.C.; Roederer, M.; Morris, L.; Seaman, M.S. Neutralization tiers of HIV-1. Curr. Opin. HIV AIDS 2018, 13, 128–136. [Google Scholar] [CrossRef]
- Pegu, A.; Hessell, A.J.; Mascola, J.R.; Haigwood, N.L. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev. 2017, 275, 296–312. [Google Scholar] [CrossRef]
- Mouquet, H. Antibody B cell responses in HIV-1 infection. Trends Immunol. 2014, 35, 549–561. [Google Scholar] [CrossRef]
- Corti, D.; Langedijk, J.P.; Hinz, A.; Seaman, M.S.; Vanzetta, F.; Fernandez-Rodriguez, B.M.; Silacci, C.; Pinna, D.; Jarrossay, D.; Balla-Jhagjhoorsingh, S.; et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 2010, 5, e8805. [Google Scholar] [CrossRef]
- Watkins, J.D.; Siddappa, N.B.; Lakhashe, S.K.; Humbert, M.; Sholukh, A.; Hemashettar, G.; Wong, Y.L.; Yoon, J.K.; Wang, W.; Novembre, F.J.; et al. An anti-HIV-1 V3 loop antibody fully protects cross-clade and elicits T-cell immunity in macaques mucosally challenged with an R5 clade C SHIV. PLoS ONE 2011, 6, e18207. [Google Scholar] [CrossRef]
- Moore, P.L.; Gray, E.S.; Sheward, D.; Madiga, M.; Ranchobe, N.; Lai, Z.; Honnen, W.J.; Nonyane, M.; Tumba, N.; Hermanus, T.; et al. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 2011, 85, 3128–3141. [Google Scholar] [CrossRef]
- Gorny, M.K.; Conley, A.J.; Karwowska, S.; Buchbinder, A.; Xu, J.Y.; Emini, E.A.; Koenig, S.; Zolla-Pazner, S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 1992, 66, 7538–7542. [Google Scholar] [CrossRef]
- Zolla-Pazner, S.; Zhong, P.; Revesz, K.; Volsky, B.; Williams, C.; Nyambi, P.; Gorny, M.K. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res. Hum. Retrovir. 2004, 20, 1254–1258. [Google Scholar] [CrossRef]
- Cerutti, N.; Loredo-Varela, J.L.; Caillat, C.; Weissenhorn, W. Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization. Curr. Opin. HIV AIDS 2017. [Google Scholar] [CrossRef]
- Kong, R.; Xu, K.; Zhou, T.; Acharya, P.; Lemmin, T.; Liu, K.; Ozorowski, G.; Soto, C.; Taft, J.D.; Bailer, R.T.; et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 2016, 352, 828–833. [Google Scholar] [CrossRef]
- Xu, K.; Acharya, P.; Kong, R.; Cheng, C.; Chuang, G.Y.; Liu, K.; Louder, M.K.; O’Dell, S.; Rawi, R.; Sastry, M.; et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 2018, 24, 857–867. [Google Scholar] [CrossRef]
- Jiang, X.; Burke, V.; Totrov, M.; Williams, C.; Cardozo, T.; Gorny, M.K.; Zolla-Pazner, S.; Kong, X.P. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 2010, 17, 955–961. [Google Scholar] [CrossRef]
- Binley, J.M.; Wrin, T.; Korber, B.; Zwick, M.B.; Wang, M.; Chappey, C.; Stiegler, G.; Kunert, R.; Zolla-Pazner, S.; Katinger, H.; et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 2004, 78, 13232–13252. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Gilbert, P.; Gurwith, M.; Heyward, W.; Martin, M.; van Griensven, F.; Hu, D.; Tappero, J.W.; Choopanya, K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 2006, 194, 1661–1671. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef]
- Corey, L.; Gilbert, P.B.; Tomaras, G.D.; Haynes, B.F.; Pantaleo, G.; Fauci, A.S. Immune correlates of vaccine protection against HIV-1 acquisition. Sci. Transl. Med. 2015, 7, 310rv317. [Google Scholar] [CrossRef]
- Gray, G.E.; Huang, Y.; Grunenberg, N.; Laher, F.; Roux, S.; Andersen-Nissen, E.; De Rosa, S.C.; Flach, B.; Randhawa, A.K.; Jensen, R.; et al. Immune correlates of the Thai RV144 HIV vaccine regimen in South Africa. Sci. transl. med. 2019, 11. [Google Scholar] [CrossRef]
- Barchichat, S.; Katz, E. Immunization of rabbits with a modified vaccinia Ankara recombinant virus bearing the HIV envelope antigen on its outer membrane. Virus Res. 2002, 90, 243–251. [Google Scholar] [CrossRef]
- Townsley, S.; Mohamed, Z.; Guo, W.; McKenna, J.; Cleveland, B.; LaBranche, C.; Beaumont, D.; Shen, X.; Yates, N.L.; Pinter, A.; et al. Induction of Heterologous Tier 2 HIV-1-Neutralizing and Cross-Reactive V1/V2-Specific Antibodies in Rabbits by Prime-Boost Immunization. J. Virol. 2016, 90, 8644–8660. [Google Scholar] [CrossRef]
- Marcelino, J.M.; Borrego, P.; Rocha, C.; Barroso, H.; Quintas, A.; Novo, C.; Taveira, N. Potent and broadly reactive HIV-2 neutralizing antibodies elicited by a vaccinia virus vector prime-C2V3C3 polypeptide boost immunization strategy. J. Virol. 2010, 84, 12429–12436. [Google Scholar] [CrossRef][Green Version]
- Narayan, K.M.; Agrawal, N.; Du, S.X.; Muranaka, J.E.; Bauer, K.; Leaman, D.P.; Phung, P.; Limoli, K.; Chen, H.; Boenig, R.I.; et al. Prime-boost immunization of rabbits with HIV-1 gp120 elicits potent neutralization activity against a primary viral isolate. PLoS ONE 2013, 8, e52732. [Google Scholar] [CrossRef]
- Quinnan, G.V., Jr.; Onabajo, O.; Zhang, P.; Yan, L.; Mattapallil, J.J.; Zhang, Z.; Dong, M.; Lu, M.; Montefiori, D.; LaBranche, C.; et al. Immunization of rabbits with highly purified, soluble, trimeric human immunodeficiency virus type 1 envelope glycoprotein induces a vigorous B cell response and broadly cross-reactive neutralization. PLoS ONE 2014, 9, e98060. [Google Scholar] [CrossRef]
- Klasse, P.J.; LaBranche, C.C.; Ketas, T.J.; Ozorowski, G.; Cupo, A.; Pugach, P.; Ringe, R.P.; Golabek, M.; van Gils, M.J.; Guttman, M.; et al. Sequential and Simultaneous Immunization of Rabbits with HIV-1 Envelope Glycoprotein SOSIP.664 Trimers from Clades A, B and C. PLoS Pathog 2016, 12, e1005864. [Google Scholar] [CrossRef]
- Zolla-Pazner, S.; Powell, R.; Yahyaei, S.; Williams, C.; Jiang, X.; Li, W.; Lu, S.; Wang, S.; Upadhyay, C.; Hioe, C.E.; et al. Rationally Designed Vaccines Targeting the V2 Region of HIV-1 gp120 Induce a Focused, Cross-Clade-Reactive, Biologically Functional Antibody Response. J. Virol. 2016, 90, 10993–11006. [Google Scholar] [CrossRef]
- de Taeye, S.W.; Moore, J.P.; Sanders, R.W. HIV-1 Envelope Trimer Design and Immunization Strategies to Induce Broadly Neutralizing Antibodies. Trends Immunol. 2016, 37, 221–232. [Google Scholar] [CrossRef]
- Dubrovskaya, V.; Tran, K.; Ozorowski, G.; Guenaga, J.; Wilson, R.; Bale, S.; Cottrell, C.A.; Turner, H.L.; Seabright, G.; O’Dell, S.; et al. Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Immunity 2019, 51, 915–929. [Google Scholar] [CrossRef]
- Francica, J.R.; Laga, R.; Lynn, G.M.; Muzikova, G.; Androvic, L.; Aussedat, B.; Walkowicz, W.E.; Padhan, K.; Ramirez-Valdez, R.A.; Parks, R.; et al. Star nanoparticles delivering HIV-1 peptide minimal immunogens elicit near-native envelope antibody responses in nonhuman primates. PLoS Biol. 2019, 17, e3000328. [Google Scholar] [CrossRef]
- Kibler, K.V.; Asbach, B.; Perdiguero, B.; Garcia-Arriaza, J.; Yates, N.L.; Parks, R.; Stanfield-Oakley, S.; Ferrari, G.; Montefiori, D.C.; Tomaras, G.D.; et al. Replication-Competent NYVAC-KC Yields Improved Immunogenicity to HIV-1 Antigens in Rhesus Macaques Compared to Nonreplicating NYVAC. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Cirelli, K.M.; Carnathan, D.G.; Nogal, B.; Martin, J.T.; Rodriguez, O.L.; Upadhyay, A.A.; Enemuo, C.A.; Gebru, E.H.; Choe, Y.; Viviano, F.; et al. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 2019, 177, 1153–1171. [Google Scholar] [CrossRef]
- Sok, D.; Le, K.M.; Vadnais, M.; Saye-Francisco, K.L.; Jardine, J.G.; Torres, J.L.; Berndsen, Z.T.; Kong, L.; Stanfield, R.; Ruiz, J.; et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 2017, 548, 108–111. [Google Scholar] [CrossRef]
- Bartolo, I.; Calado, R.; Borrego, P.; Leitner, T.; Taveira, N. Rare HIV-1 Subtype J Genomes and a New H/U/CRF02_AG Recombinant Genome Suggests an Ancient Origin of HIV-1 in Angola. AIDS Res. Hum. Retrovir. 2016, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Borrego, P.; Calado, R.; Marcelino, J.M.; Pereira, P.; Quintas, A.; Barroso, H.; Taveira, N. An ancestral HIV-2/simian immunodeficiency virus peptide with potent HIV-1 and HIV-2 fusion inhibitor activity. AIDS 2013, 27, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Borrego, P.; Calado, R.; Marcelino, J.M.; Bartolo, I.; Rocha, C.; Cavaco-Silva, P.; Doroana, M.; Antunes, F.; Maltez, F.; Caixas, U.; et al. Baseline susceptibility of primary HIV-2 to entry inhibitors. Antivir. Ther. 2012, 17, 565–570. [Google Scholar] [CrossRef]
- Lengauer, T.; Sander, O.; Sierra, S.; Thielen, A.; Kaiser, R. Bioinformatics prediction of HIV coreceptor usage. Nat. Biotechnol. 2007, 25, 1407–1410. [Google Scholar] [CrossRef]
- Davison, A.J.; Moss, B. New vaccinia virus recombination plasmids incorporating a synthetic late promoter for high level expression of foreign proteins. Nucleic Acids Res. 1990, 18, 4285–4286. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Rose, N.F.; Roberts, A.; Buonocore, L.; Rose, J.K. Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J. Virol. 2000, 74, 10903–10910. [Google Scholar] [CrossRef]
- Arthos, J. Purification of HIV Envelope Protein. Available online: http://www.aidsreagent.org/pdf_images/4126_001.pdf (accessed on 5 July 2018).
- Laboratory, M. Protocol for Heat-Inactivation of Serum and Plasma Samples. Available online: http://www.hiv.lanl.gov/content/nab-reference-strains/html/Protocol-for-Heat-Inactivation-of-Serum-and-Plasma-Samples_Jan2016.pdf (accessed on 19 May 2018).
- deCamp, A.; Hraber, P.; Bailer, R.T.; Seaman, M.S.; Ochsenbauer, C.; Kappes, J.; Gottardo, R.; Edlefsen, P.; Self, S.; Tang, H.; et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2014, 88, 2489–2507. [Google Scholar] [CrossRef]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Kilby, J.M.; Saag, M.S.; et al. Antibody neutralization and escape by HIV-1. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef]
- Sarzotti-Kelsoe, M.; Bailer, R.T.; Turk, E.; Lin, C.L.; Bilska, M.; Greene, K.M.; Gao, H.; Todd, C.A.; Ozaki, D.A.; Seaman, M.S.; et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 2014, 409, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Eroshkin, A.M.; LeBlanc, A.; Weekes, D.; Post, K.; Li, Z.; Rajput, A.; Butera, S.T.; Burton, D.R.; Godzik, A. bNAber: Database of broadly neutralizing HIV antibodies. Nucleic Acids Res 2014, 42, D1133–D1139. [Google Scholar] [CrossRef] [PubMed]
- Finzi, A.; Pacheco, B.; Zeng, X.; Kwon, Y.D.; Kwong, P.D.; Sodroski, J. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods 2010, 168, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.M.; Liao, H.X.; Tomaras, G.D.; Bonsignori, M.; Tsao, C.Y.; Hwang, K.K.; Chen, H.; Lloyd, K.E.; Bowman, C.; Sutherland, L.; et al. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J. virol. 2013, 87, 1554–1568. [Google Scholar] [CrossRef] [PubMed]
- van den Kerkhof, T.L.; van Gils, M.J.; Boeser-Nunnink, B.D.; Burger, J.A.; Schuitemaker, H.; Sanders, R.W. Probability of N332 glycan occupancy on HIV-1 gp120 modulates sensitivity to broadly neutralizing antibodies. AIDS 2016, 30, 2179–2184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bartolo, I.; Casanovas, J.; Bastos, R.; Rocha, C.; Abecasis, A.B.; Folgosa, E.; Mondlane, J.; Manuel, R.; Taveira, N. HIV-1 genetic diversity and transmitted drug resistance in health care settings in Maputo, Mozambique. J. Acquir. Immune Defic. Syndr. 2009, 51, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Thippeshappa, R.; Tian, B.; Cleveland, B.; Guo, W.; Polacino, P.; Hu, S.L. Oral Immunization with Recombinant Vaccinia Virus Prime and Intramuscular Protein Boost Provides Protection against Intrarectal Simian-Human Immunodeficiency Virus Challenge in Macaques. Clin Vaccine Immunol 2015, 23, 204–212. [Google Scholar] [CrossRef]
- Kibler, K.V.; Asbach, B.; Perdiguero, B.; Garcia-Arriaza, J.; Yates, N.L.; Parks, R.; Stanfield-Oakley, S.; Ferrari, G.; Montefiori, D.C.; Tomaras, G.D.; et al. Correction for Kibler et al., "Replication-Competent NYVAC-KC Yields Improved Immunogenicity to HIV-1 Antigens in Rhesus Macaques Compared to Nonreplicating NYVAC". J. virol. 2019, 93. [Google Scholar] [CrossRef]
- Bartolo, I.; Rocha, C.; Bartolomeu, J.; Gama, A.; Marcelino, R.; Fonseca, M.; Mendes, A.; Epalanga, M.; Silva, P.C.; Taveira, N. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: New insights into the origins of the AIDS pandemic. Infect. Genet. Evol. 2009, 9, 672–682. [Google Scholar] [CrossRef]
- Pineda-Pena, A.C.; Varanda, J.; Sousa, J.D.; Theys, K.; Bartolo, I.; Leitner, T.; Taveira, N.; Vandamme, A.M.; Abecasis, A.B. On the contribution of Angola to the initial spread of HIV-1. Infect. Genet. Evol. 2016, 46, 219–222. [Google Scholar] [CrossRef]
- Barroso, H.; Borrego, P.; Bartolo, I.; Marcelino, J.M.; Familia, C.; Quintas, A.; Taveira, N. Evolutionary and structural features of the C2, V3 and C3 envelope regions underlying the differences in HIV-1 and HIV-2 biology and infection. PLoS ONE 2011, 6, e14548. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Tomaka, F.L.; Wegmann, F.; Stieh, D.J.; Alter, G.; Robb, M.L.; Michael, N.L.; Peter, L.; Nkolola, J.P.; Borducchi, E.N.; et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet 2018, 392, 232–243. [Google Scholar] [CrossRef]
- Kim, M.; Qiao, Z.S.; Montefiori, D.C.; Haynes, B.F.; Reinherz, E.L.; Liao, H.X. Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retrovir. 2005, 21, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Svehla, K.; Mathy, N.L.; Voss, G.; Mascola, J.R.; Wyatt, R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 2006, 80, 1414–1426. [Google Scholar] [CrossRef]
- Beddows, S.; Franti, M.; Dey, A.K.; Kirschner, M.; Iyer, S.P.; Fisch, D.C.; Ketas, T.; Yuste, E.; Desrosiers, R.C.; Klasse, P.J.; et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 2007, 360, 329–340. [Google Scholar] [CrossRef]
- Flynn, N.M.; Forthal, D.N.; Harro, C.D.; Judson, F.N.; Mayer, K.H.; Para, M.F. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 2005, 191, 654–665. [Google Scholar] [CrossRef]
- Buchbinder, S.P.; Mehrotra, D.V.; Duerr, A.; Fitzgerald, D.W.; Mogg, R.; Li, D.; Gilbert, P.B.; Lama, J.R.; Marmor, M.; Del Rio, C.; et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008, 372, 1881–1893. [Google Scholar] [CrossRef]
- Nkolola, J.P.; Peng, H.; Settembre, E.C.; Freeman, M.; Grandpre, L.E.; Devoy, C.; Lynch, D.M.; La Porte, A.; Simmons, N.L.; Bradley, R.; et al. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J. Virol. 2010, 84, 3270–3279. [Google Scholar] [CrossRef]
- Yusim, K.D.B.; Koup, R.; Korber, B.T.M.; de Boer, R.; Moore, J.P.; Watkins, D.I.; Brander, C.; Haynes, B.F.; Walker, B.D. HIV Molecular Immunology; Los Alamos National Laboratory: Los Alamos, NM, USA, 2015.
- Murphy, G.; Belda, F.J.; Pau, C.P.; Clewley, J.P.; Parry, J.V. Discrimination of subtype B and non-subtype B strains of human immunodeficiency virus type 1 by serotyping: Correlation with genotyping. J Clin Microbiol 1999, 37, 1356–1360. [Google Scholar] [CrossRef]
- Burke, V.; Williams, C.; Sukumaran, M.; Kim, S.S.; Li, H.; Wang, X.H.; Gorny, M.K.; Zolla-Pazner, S.; Kong, X.P. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: Implications for design of V3-based immunogens. Structure 2009, 17, 1538–1546. [Google Scholar] [CrossRef]
- Cardozo, T.; Swetnam, J.; Pinter, A.; Krachmarov, C.; Nadas, A.; Almond, D.; Zolla-Pazner, S. Worldwide distribution of HIV type 1 epitopes recognized by human anti-V3 monoclonal antibodies. AIDS Res. Hum. Retrovir. 2009, 25, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Doores, K.J.; Kong, L.; Krumm, S.A.; Le, K.M.; Sok, D.; Laserson, U.; Garces, F.; Poignard, P.; Wilson, I.A.; Burton, D.R. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 2015, 89, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Sok, D.; Doores, K.J.; Briney, B.; Le, K.M.; Saye-Francisco, K.L.; Ramos, A.; Kulp, D.W.; Julien, J.P.; Menis, S.; Wickramasinghe, L.; et al. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci. Transl. Med. 2014, 6, 236ra263. [Google Scholar] [CrossRef] [PubMed]
- Gavel, Y.; von Heijne, G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. 1990, 3, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Mellquist, J.L.; Kasturi, L.; Spitalnik, S.L.; Shakin-Eshleman, S.H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837. [Google Scholar] [CrossRef] [PubMed]







| Groups | Mice ID | HIV-1 Env Clade | Priming | Boost I | Boost II | Boost III |
|---|---|---|---|---|---|---|
| Day 1 | Day 15 | Day 30 | Day 45 | |||
| 1 | M1-M2 | - | - | - | - | - |
| 2 | M3-M5 | - | VVWR | - | - | - |
| 3 | M6-M8 | C | VVgp120t-C | C2V3C3-Ca | C2V3C3-Ca | Sgp120t-C |
| 4 | M9-M11 | C | VVgp120t-C | C2V3C3-Cb | C2V3C3-Cb | Sgp120t-C |
| 5 | M12-M16 | CRF02_AG | VVgp120t-AG | C2V3C3-AG | C2V3C3-AG | Sgp120t-AG |
| 6 | M17-M20 | J | VVgp120t-J | C2V3C3-J | C2V3C3-J | Sgp120t-J |
| Groups | Mice ID | HIV-1 Env Clade | Priming | Boost I | Boost II | Boost III |
|---|---|---|---|---|---|---|
| Day 1 | Day 15 | Day 30 | Day 45 | |||
| 1 | M1-M2 | - | - | - | - | - |
| 2 | M3-M4 | - | VVWR | SWR | SWR | SWR |
| 3 | M5-M8 | C | VVgp120t-C | C2V3C3-C | C2V3C3-C | Sgp120t-C |
| 3A | M9-M11 | C | VVgp120t-C | Sgp120C | Sgp120C | Sgp120t-C |
| 3B | M12-M13 | C | Sgp120t-C | C2V3C3-C | C2V3C3-C | Sgp120t-C |
| 4 | M14-M17 | CRF02_AG | VVgp120t-AG | C2V3C3-AG | C2V3C3-AG | Sgp120t-AG |
| 4A | M18-M20 | CRF02_AG | VVgp120t-AG | Sgp120t-AG | Sgp120t-AG | Sgp120t-AG |
| 4B | M21-M22 | CRF02_AG | Sgp120t-AG | C2V3C3-AG | C2V3C3-AG | Sgp120t-AG |
| 5 | M23-M26 | B | VVgp120t-B | C2V3C3-C | C2V3C3-C | Sgp120t-B |
| 5A | M27-M28 | B | VVgp120t-B | Sgp120t-B | Sgp120t-B | Sgp120t-B |
| 5B | M29-M30 | B | Sgp120t-B | C2V3C3-C | C2V3C3-C | Sgp120t-B |
| Groups | ID | Blood Collection | Priming | Blood Collection | Boost I | Blood Collection | Boost II | Blood Collection | Blood Collection | Boost III | Boost IV | Blood Collection |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 (T0) | Day 1 | Day 35 (T1) | Day 35 | Day 63 (T2) | Day 63 | Day 99 (T3) | Day 121 (T4) | Day 121 | Day 150 | Day 154 (T5) | ||
| 1 | R1 | ● | - | ● | - | ● | - | ● | ● | - | - | ● |
| R2 | ● | PBS | ● | PBS | ● | PBS | ● | ● | PBS | - | ● | |
| 2 | R3 | ● | VVWR | ● | SWR | ● | SWR | ● | ● | SWR | - | ● |
| R4 | ● | VVWR | ● | SWR | ● | SWR | ● | ● | SWR | - | ● | |
| 3 | R5 | ● | VVgp120t-AG | ● | Sgp120t-AG | ● | Sgp120t-AG | ● | ● | Sgp120t-AG | - | ● |
| R6 | ● | VVgp120t-AG | ● | Sgp120t-AG | ● | Sgp120t-AG | ● | ● | Sgp120t-AG | Sgp120t-AG | ● | |
| R7 | ● | VVgp120t-AG | ● | Sgp120t-AG | ● | Sgp120t-AG | ● | ● | Sgp120t-AG | - | ● | |
| R8 | ● | VVgp120t-AG | ● | Sgp120t-AG | ● | Sgp120t-AG | ● | ● | Sgp120t-AG | Sgp120t-AG | ● |
| Immunization Scheme | Mice ID | Group | Immunogen Clade | Tier 1 Isolate | Tier 2 Pseudovirus Panel | Primary Isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL4.3 | 398F1 | X2278 | TRO11 | 25710 | CE0217 | CE1176 | X1632 | CNE55 | CH119 | BJOX2000 | 246F3 | 01PTCJN | 93HDC250 | ||||
| B | A | B | B | C | C | C | G | CRF07_BC | CRF07_BC | CRF07_BC | AC | CRF02_AG | J | ||||
| Mock control | M1-M2 | 1 | _ | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| Vaccinia control (VVWR+SWR) | M3-M5 | 2 | _ | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| VVgp120t+C2V3C3+C2V3C3+Sgp120t | M6 | 3 | C | 58 | 69 | <30 | <30 | 59 | 35 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| M7 | ND | 61 | <30 | <30 | ND | 47 | <30 | <30 | 41 | <30 | <30 | <30 | <30 | <30 | |||
| M8 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | 50 | |||
| M9 | 4 | C | ND | 47 | ND | 31 | ND | ND | <30 | ND | ND | ND | ND | ND | ND | ND | |
| M10 | 49 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M11 | ND | <30 | <30 | <30 | ND | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M12 | 5 | CRF02_AG | <30 | 37 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |
| M13 | ND | 76 | ND | <30 | ND | 56 | <30 | 50 | 57 | ND | 47 | <30 | 49 | 55 | |||
| M15 | ND | <30 | <30 | <30 | ND | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M16 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M17 | 6 | J | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | ND | ND | <30 | <30 | ND | <30 | |
| M18 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M19 | 73 | <30 | <30 | <30 | 54 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| M20 | ND | <30 | <30 | <30 | ND | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |||
| Control of neutralization* | 95 | 78 | 48 | 69 | 92 | 63 | 91 | 90 | 67 | 75 | 78 | 75 | 51 | 87 | |||
| Immunization Scheme | Groups | Immunogen Clade | Tier 1 Isolate | Tier 2 Pseudovirus Panel | Primary Isolates | VSV | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SG3.1 | 398F1 | X2278 | TRO11 | 25710 | CE0217 | CE1176 | X1632 | CNE55 | CH119 | BJOX2000 | 246F3 | 01PTCJN | 93HDC250 | 93HDC253 | ||||
| B | A | B | B | C | C | C | G | CRF07_BC | CRF07_BC | CRF07_BC | AC | CRF02_AG | J | J | ||||
| Mock control | 1 | _ | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| Vaccinia control (VVWR+SWR) | 2 | _ | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| VVgp120t+C2V3C3+C2V3C3+Sgp120t | 3 | C | <30 | <30 | 40 | 52 | <30 | <30 | <30 | <30 | <30 | 34 | <30 | <30 | 54 | <30 | <30 | <30 |
| VVgp120t+Sgp120t+Sgp120t+gp120t | 3A | <30 | <30 | 42 | <30 | <30 | <30 | <30 | <30 | <30 | ND | <30 | <30 | 46 | ND | ND | <30 | |
| Sgp120t+C2V3C3+C2V3C3+Sgp120t | 3B | <30 | <30 | 55 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |
| VVgp120t+C2V3C3+Sgp120t | 4 | CRF02_AG | <30 | <30 | 53 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| VVgp120t+Sgp120t+Sgp120t+Sgp120t | 4A | 56 | <30 | 52 | 44 | <30 | <30 | <30 | <30 | 36 | 49 | <30 | <30 | 70 | <30 | 50 | <30 | |
| Sgp120t+C2V3C3+C2V3C3+Sgp120t | 4B | 44 | 38 | 51 | <30 | <30 | <30 | <30 | <30 | <30 | ND | <30 | <30 | ND | ND | ND | <30 | |
| VVgp120t+C2V3C3+Sgp120t | 5 | B+C | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | 48 | <30 | <30 | 58 | <30 | <30 | <30 |
| VVgp120t+Sgp120t+Sgp120t+gp120t | 5A | <30 | 43 | <30 | ND | <30 | <30 | <30 | <30 | 35 | ND | <30 | <30 | 44 | ND | ND | <30 | |
| Sgp120t+C2V3C3+C2V3C3+Sgp120t | 5B | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | |
| Control of neutralization* | 99 | 67 | 93 | 69 | 92 | 94 | 91 | 80 | 95 | 57 | 78 | 95 | 51 | 38 | 75 | <30 | ||
| Immunization Scheme and Controls | Animal ID | HIV-1 Pseudoviruses | Primary Isolates | Specificity Controls | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tier 1 | Tier 2 | HIV-1 | HIV-2 Isolates | VSV | ||||||||||||||||||
| NL 4.3 | SG 3.1 | PCE 0217 | PCNE8 | PCH 119 | TRO 11 | CNE 55 | CE 1176 | 246 F3 | BJOX 2000 | X 1632 | X 2278 | 398 F1 | 25 710 | 01PT CJN | 93HD C252 | 93HD C253 | 93HD C249 | HCC 19.03 | AUC | |||
| B | B | C | 01 _AE | 07_BC | B | 07 _BC | C | AC | 07 _BC | G | B | A | C | 02 _AG | U | J | U | A | A | |||
| Mock | R1-R2 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| Vaccinia (VVWR+SWR) | R3-R4 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| VV gp120t- AG+Sgp120t-AG | R5-R6 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | <30 |
| R7 | <30 | 36 | <30 | <30 | <30 | <30 | <30 | <30 | 39 | <30 | 45 | <30 | <30 | <30 | 32 | 33 | <30 | <30 | <30 | <30 | <30 | |
| R8 | 58 (20) | 63 (20) | 30 | 90 (20) | 36 | 32 | 58 (20) | 54 (20) | 80 (20) | 61 (20) | 77 (20) | 79 (20) | 66 (20) | 86 (80) | 71 (40) | 91 (40) | 65 (20) | 75 (20) | <30 | <30 | <30 | |
| Neutralization controls | bNAbs | 94 | 95 | 92 | 70 | 85 | 82 | 62 | <30 | 66 | <30 | <30 | 93 | 79 | <30 | 90 | 85 | 84 | <30 | <30 | <30 | <30 |
| HIV-1* | 89 | 82 | 96 | 100 | 98 | 97 | 95 | 95 | 86 | 79 | 81 | 92 | 99 | 99 | 97 | 91 | 92 | 96 | <30 | <30 | <30 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calado, R.; Duarte, J.; Borrego, P.; Marcelino, J.M.; Bártolo, I.; Martin, F.; Figueiredo, I.; Almeida, S.; Graça, L.; Vítor, J.; et al. A Prime-Boost Immunization Strategy with Vaccinia Virus Expressing Novel gp120 Envelope Glycoprotein from a CRF02_AG Isolate Elicits Cross-Clade Tier 2 HIV-1 Neutralizing Antibodies. Vaccines 2020, 8, 171. https://doi.org/10.3390/vaccines8020171
Calado R, Duarte J, Borrego P, Marcelino JM, Bártolo I, Martin F, Figueiredo I, Almeida S, Graça L, Vítor J, et al. A Prime-Boost Immunization Strategy with Vaccinia Virus Expressing Novel gp120 Envelope Glycoprotein from a CRF02_AG Isolate Elicits Cross-Clade Tier 2 HIV-1 Neutralizing Antibodies. Vaccines. 2020; 8(2):171. https://doi.org/10.3390/vaccines8020171
Chicago/Turabian StyleCalado, Rita, Joana Duarte, Pedro Borrego, José Maria Marcelino, Inês Bártolo, Francisco Martin, Inês Figueiredo, Silvia Almeida, Luís Graça, Jorge Vítor, and et al. 2020. "A Prime-Boost Immunization Strategy with Vaccinia Virus Expressing Novel gp120 Envelope Glycoprotein from a CRF02_AG Isolate Elicits Cross-Clade Tier 2 HIV-1 Neutralizing Antibodies" Vaccines 8, no. 2: 171. https://doi.org/10.3390/vaccines8020171
APA StyleCalado, R., Duarte, J., Borrego, P., Marcelino, J. M., Bártolo, I., Martin, F., Figueiredo, I., Almeida, S., Graça, L., Vítor, J., Aires da Silva, F., Dias, I., Carrapiço, B., & Taveira, N. (2020). A Prime-Boost Immunization Strategy with Vaccinia Virus Expressing Novel gp120 Envelope Glycoprotein from a CRF02_AG Isolate Elicits Cross-Clade Tier 2 HIV-1 Neutralizing Antibodies. Vaccines, 8(2), 171. https://doi.org/10.3390/vaccines8020171