Disruption of the cpsE and endA Genes Attenuates Streptococcus pneumoniae Virulence: Towards the Development of a Live Attenuated Vaccine Candidate
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Construction of cpsE::tetL and endA::aphA-3 Knockout Mutants
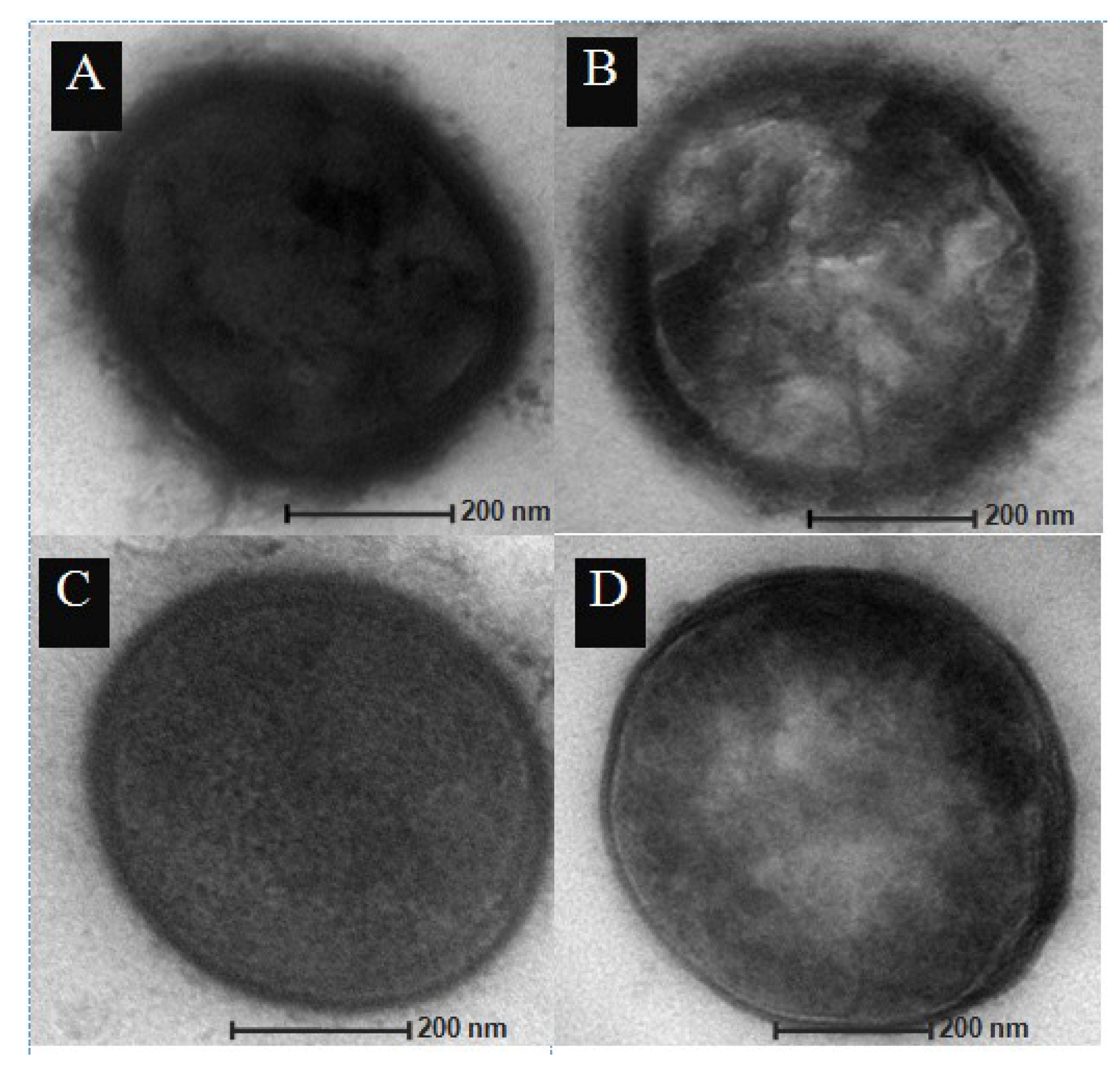
2.3. Transmission Electron Microscopy
2.4. In Vitro Studies
2.5. In Vivo Studies
3. Results
3.1. Disruption of the cpsE Gene Led to Loss of the Capsule in S. Pneumoniae
3.2. In Vitro Growth Characteristics
3.3. In Vitro Biofilm Formation
3.4. Density and Duration of Nasopharyngeal (NP) Colonisation in Mice
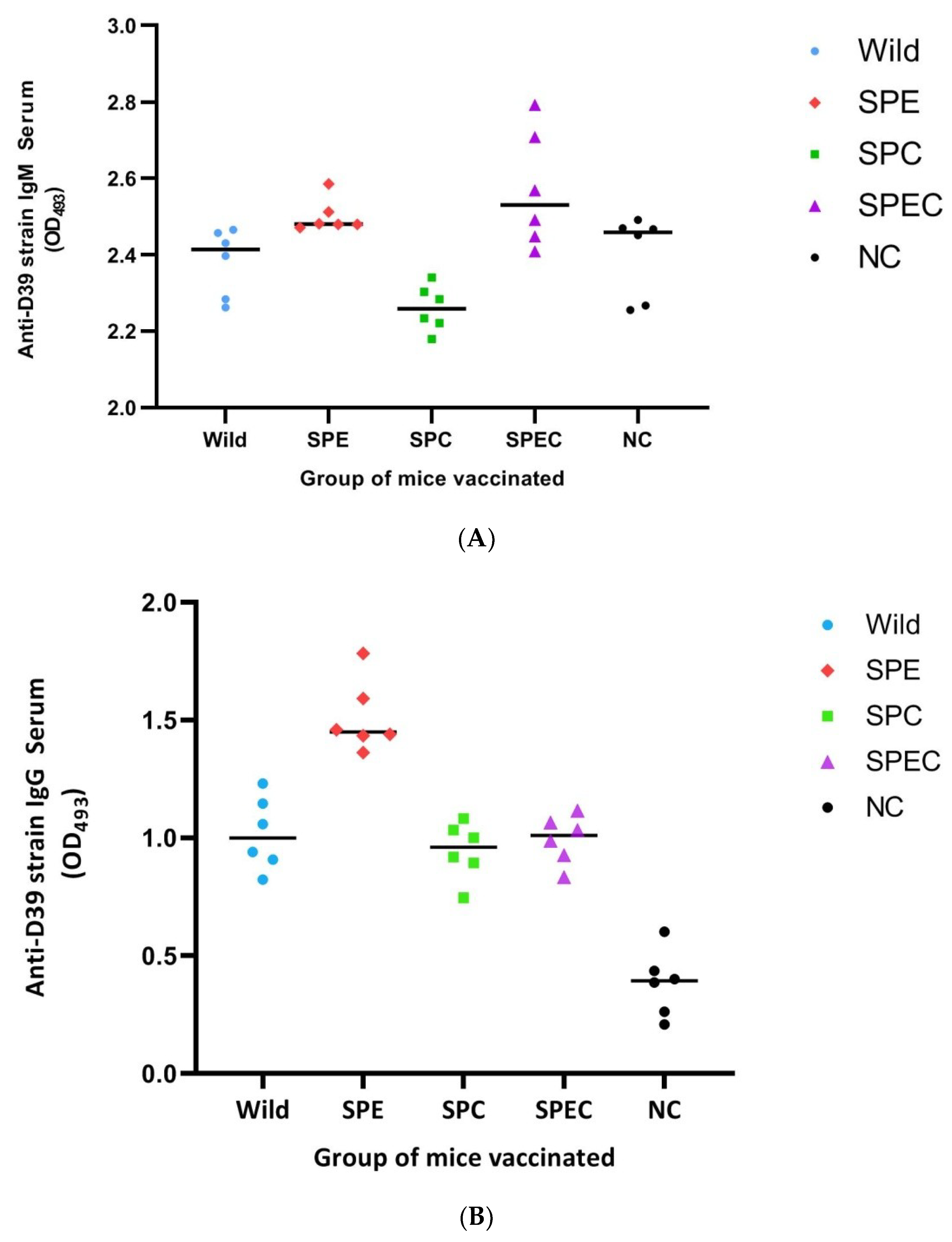
3.5. Effect of Immunisation with the Gene-Disrupted Mutant Strains on Local and Systemic Antibody Titres
3.6. Survival Analysis
3.7. Attenuation of Virulence
4. Discussion
4.1. Capsule Mutations Affected the Growth of S. Pneumoniae
4.2. Biofilm Profiles of the Candidate Vaccine Strains
4.3. Colonisation Potential of the Pneumococcal Gene-Knockout Strains
4.4. Antibody Response to Candidate Vaccine Strains
4.5. Immune Protection Conferred by Gene-Knockout Strains
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Troeger, C.; Blacker, B.; Khalil, I.; Rao, P.; Cao, J.; Zimsen, S.; Albertson, S.; Deshpande, A.; Farag, T.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- Wahl, B.; O’Brien, K.L.; Greenbaum, A.; Majumder, A.; Liu, L.; Chu, Y.; Luksic, I.; Nair, H.; McAllister, D.A.; Campbell, H.; et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: Global, regional, and national estimates for 2000–15. Lancet Glob. Health 2018, 6, e744–e757. [Google Scholar] [CrossRef]
- O’Brien, K.L.; Wolfson, L.J.; Watt, J.P.; Henkle, E.; Deloria-Knoll, M.; McCall, N.; Lee, E.; Mulholland, K.; Levine, O.S.; Cherian, T.; et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet 2009, 374, 893–902. [Google Scholar] [CrossRef]
- Bryce, J.; Boschi-Pinto, C.; Shibuya, K.; Black, R.E.; WHO Child Health Epidemiology Reference Group. WHO estimates of the causes of death in children. Lancet 2005, 365, 1147–1152. [Google Scholar] [CrossRef]
- Mook-Kanamori, B.B.; Geldhoff, M.; van der Poll, T.; van de Beek, D. Pathogenesis and pathophysiology of pneumococcal meningitis. Clin. Microbiol. Rev. 2011, 24, 557–591. [Google Scholar] [CrossRef] [PubMed]
- Malley, R. Antibody and cell-mediated immunity to Streptococcus pneumoniae: Implications for vaccine development. J. Mol. Med (Berl.) 2010, 88, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Geno, K.A.; Gilbert, G.L.; Song, J.Y.; Skovsted, I.C.; Klugman, K.P.; Jones, C.; Konradsen, H.B.; Nahm, M.H. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin. Microbiol. Rev. 2015, 28, 871–899. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Yamagishi, Y.; Hirai, J.; Sakanashi, D.; Suematsu, H.; Nishiyama, N.; Koizumi, Y.; Mikamo, H. Invasive pneumococcal disease caused by mucoid serotype 3 Streptococcus pneumoniae: A case report and literature review. BMC Res. Notes 2017, 10, 21. [Google Scholar] [CrossRef][Green Version]
- Jauneikaite, E.; Jefferies, J.M.; Hibberd, M.L.; Clarke, S.C. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: A review. Vaccine 2012, 30, 3503–3514. [Google Scholar] [CrossRef]
- VIEW-hub Global Vaccine Introduction Report, December 2018. Available online: www.jhsph.edu/ivac/view-hub. (accessed on 14 June 2019).
- Tricarico, S.; McNeil, H.C.; Cleary, D.W.; Head, M.G.; Lim, V.; Yap, I.K.S.; Wie, C.C.; Tan, C.S.; Norazmi, M.N.; Aziah, I.; et al. Pneumococcal conjugate vaccine implementation in middle-income countries. Pneumonia (Nathan) 2017, 9, 6. [Google Scholar] [CrossRef]
- Hicks, L.A.; Harrison, L.H.; Flannery, B.; Hadler, J.L.; Schaffner, W.; Craig, A.S.; Jackson, D.; Thomas, A.; Beall, B.; Lynfield, R.; et al. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 2007, 196, 1346–1354. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.; Andrews, N.J.; Waight, P.A.; Slack, M.P.; George, R.C. Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: An observational cohort study. Lancet Infect. Dis. 2011, 11, 760–768. [Google Scholar] [CrossRef]
- Henriques-Normark, B.; Tuomanen, E.I. The pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Gladstone, R.A.; Jefferies, J.M.; Tocheva, A.S.; Beard, K.R.; Garley, D.; Chong, W.W.; Bentley, S.D.; Faust, S.N.; Clarke, S.C. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine 2015, 33, 2015–2021. [Google Scholar] [CrossRef]
- Janapatla, R.P.; Su, L.H.; Chen, H.H.; Chang, H.J.; Tsai, T.C.; Chen, P.Y.; Chen, C.L.; Chiu, C.H. Epidemiology of culture-confirmed infections of Streptococcus pneumoniae (2012–2015) and nasopharyngeal carriage in children and households in Taiwan (2014–2015). J. Med. Microbiol. 2017, 66, 729–736. [Google Scholar] [CrossRef]
- Vestrheim, D.F.; Steinbakk, M.; Aaberge, I.S.; Caugant, D.A. Postvaccination increase in serotype 19A pneumococcal disease in Norway is driven by expansion of penicillin-susceptible strains of the ST199 complex. Clin. Vaccine. Immunol. 2012, 19, 443–445. [Google Scholar] [CrossRef]
- Plotkin, S.A.; Orenstein, W.A.; Offit, P.A.; Edwards, K.M. Plotkin’s Vaccines; Elsevier: Philadelphia, PA, USA, 2018; pp. 18–26. [Google Scholar]
- Lockhart, S. Conjugate vaccines. Expert. Rev. Vaccine. 2003, 2, 633–648. [Google Scholar] [CrossRef]
- Guevara, M.; Barricarte, A.; Torroba, L.; Herranz, M.; Gil-Setas, A.; Gil, F.; Bernaola, E.; Ezpeleta, C.; Castilla, J.; Working Group for Surveillance of the Pneumococcal Invasive Disease in Navarra. Direct, indirect and total effects of 13-valent pneumococcal conjugate vaccination on invasive pneumococcal disease in children in Navarra, Spain, 2001 to 2014: Cohort and case-control study. Euro. Surveill. 2016, 21. [Google Scholar] [CrossRef]
- Esposito, S.; Principi, N.; Group, E.V.S. Direct and indirect effects of the 13-valent pneumococcal conjugate vaccine administered to infants and young children. Future Microbiol. 2015, 10, 1599–1607. [Google Scholar] [CrossRef]
- Vetter, V.; Denizer, G.; Friedland, L.R.; Krishnan, J.; Shapiro, M. Understanding modern-day vaccines: What you need to know. An. Med. 2018, 50, 110–120. [Google Scholar] [CrossRef]
- Moffitt, K.L.; Yadav, P.; Weinberger, D.M.; Anderson, P.W.; Malley, R. Broad antibody and T cell reactivity induced by a pneumococcal whole-cell vaccine. Vaccine 2012, 30, 4316–4322. [Google Scholar] [CrossRef] [PubMed]
- Roche, A.M.; King, S.J.; Weiser, J.N. Live attenuated Streptococcus pneumoniae strains induce serotype-independent mucosal and systemic protection in mice. Infect. Immun. 2007, 75, 2469–2475. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, T.O.; Hinds, J.; Gould, K.A.; Wuthrich, D.; Bruggmann, R.; Kuffer, M.; Muhlemann, K.; Hilty, M.; Hathaway, L.J. A point mutation in cpsE renders Streptococcus pneumoniae nonencapsulated and enhances its growth, adherence and competence. BMC Microbiol. 2014, 14, 210. [Google Scholar] [CrossRef]
- Beiter, K.; Wartha, F.; Albiger, B.; Normark, S.; Zychlinsky, A.; Henriques-Normark, B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 2006, 16, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.J.; Kireev, D.; Moon, A.F.; Midon, M.; Janzen, W.P.; Pingoud, A.; Pedersen, L.C.; Singleton, S.F. Inhibitors of Streptococcus pneumoniae surface endonuclease EndA discovered by high-throughput screening using a PicoGreen fluorescence assay. J. Biomol. Screen 2013, 18, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Rosch, J.W.; Iverson, A.R.; Humann, J.; Mann, B.; Gao, G.; Vogel, P.; Mina, M.; Murrah, K.A.; Perez, A.C.; Edward Swords, W.; et al. A live-attenuated pneumococcal vaccine elicits CD4+ T-cell dependent class switching and provides serotype independent protection against acute otitis media. EMBO Mol. Med. 2014, 6, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, H.; Liu, Y.; Wang, Y.; Zeng, L.; Wu, K.; Wang, J.; Ma, F.; Xu, W.; Yin, Y.; et al. Mucosal immunization with the live attenuated vaccine SPY1 induces humoral and Th2-Th17-regulatory T cell cellular immunity and protects against pneumococcal infection. Infect. Immun. 2015, 83, 90–100. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, J.; Wu, Y.; Wang, H.; Wang, J.; Qiu, Y.; Mo, Y.; He, Y.; Zhang, X.; Yin, Y.; et al. Streptococcus pneumoniae Attenuated Strain SPY1 with an Artificial Mineral Shell Induces Humoral and Th17 Cellular Immunity and Protects Mice against Pneumococcal Infection. Front. Immunol. 2017, 8, 1983. [Google Scholar] [CrossRef]
- Nieto, C.; Espinosa, M. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 2003, 49, 281–285. [Google Scholar] [CrossRef]
- Thanassi, J.A.; Hartman-Neumann, S.L.; Dougherty, T.J.; Dougherty, B.A.; Pucci, M.J. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic. Acids. Res. 2002, 30, 3152–3162. [Google Scholar] [CrossRef]
- Hammerschmidt, S.; Wolff, S.; Hocke, A.; Rosseau, S.; Muller, E.; Rohde, M. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 2005, 73, 4653–4667. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Chae, S.W.; Song, J.J. In VitroStreptococcus pneumoniae Biofilm Formation and In Vivo Middle Ear Mucosal Biofilm in a Rat Model of Acute Otitis Induced by S. pneumoniae. Clin. Exp. Otorhinolaryngol. 2012, 5, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Cohen, J.M.; Chimalapati, S.; de Vogel, C.; van Belkum, A.; Baxendale, H.E.; Brown, J.S. Contributions of capsule, lipoproteins and duration of colonisation towards the protective immunity of prior Streptococcus pneumoniae nasopharyngeal colonisation. Vaccine 2012, 30, 4453–4459. [Google Scholar] [CrossRef]
- Choi, S.Y.; Tran, T.D.; Briles, D.E.; Rhee, D.K. Inactivated pep27 mutant as an effective mucosal vaccine against a secondary lethal pneumococcal challenge in mice. Clin. Exp. Vaccine. Res. 2013, 2, 58–65. [Google Scholar] [CrossRef][Green Version]
- Avci, F.Y.; Kasper, D.L. How bacterial carbohydrates influence the adaptive immune system. Annu. Rev. Immunol. 2010, 28, 107–130. [Google Scholar] [CrossRef]
- Brodsky, I.E.; Medzhitov, R. Targeting of immune signalling networks by bacterial pathogens. Nat. Cell Biol. 2009, 11, 521–526. [Google Scholar] [CrossRef]
- Keller, L.E.; Robinson, D.A.; McDaniel, L.S. Nonencapsulated Streptococcus pneumoniae: Emergence and Pathogenesis. MBio 2016, 7, e01792. [Google Scholar] [CrossRef]
- Iannelli, F.; Pearce, B.J.; Pozzi, G. The type 2 capsule locus of Streptococcus pneumoniae. J. Bacteriol. 1999, 181, 2652–2654. [Google Scholar] [CrossRef]
- Morona, J.K.; Miller, D.C.; Morona, R.; Paton, J.C. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 2004, 189, 1905–1913. [Google Scholar] [CrossRef]
- Cartee, R.T.; Forsee, W.T.; Bender, M.H.; Ambrose, K.D.; Yother, J. CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation. J. Bacteriol. 2005, 187, 7425–7433. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, L.; Boumedienne, M.; Saksouk, N.; Geiselmann, J.; Geremia, R.A. The glucosyl-1-phosphate transferase WchA (Cap8E) primes the capsular polysaccharide repeat unit biosynthesis of Streptococcus pneumoniae serotype 8. Biochem. Biophys. Res. Commun. 2005, 327, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Guidolin, A.; Morona, J.K.; Morona, R.; Hansman, D.; Paton, J.C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect. Immun. 1994, 62, 5384–5396. [Google Scholar] [CrossRef] [PubMed]
- Kolkman, M.A.; Morrison, D.A.; Van Der Zeijst, B.A.; Nuijten, P.J. The capsule polysaccharide synthesis locus of streptococcus pneumoniae serotype 14: Identification of the glycosyl transferase gene cps14E. J. Bacteriol. 1996, 178, 3736–3741. [Google Scholar] [CrossRef][Green Version]
- Hardy, G.G.; Caimano, M.J.; Yother, J. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 2000, 182, 1854–1863. [Google Scholar] [CrossRef]
- Kurola, P.; Erkkila, L.; Kaijalainen, T.; Palmu, A.A.; Hausdorff, W.P.; Poolman, J.; Jokinen, J.; Kilpi, T.M.; Leinonen, M.; Saukkoriipi, A. Presence of capsular locus genes in immunochemically identified encapsulated and unencapsulated Streptococcus pneumoniae sputum isolates obtained from elderly patients with acute lower respiratory tract infection. J. Med. Microbiol. 2010, 59, 1140–1145. [Google Scholar] [CrossRef][Green Version]
- Fox, C.B.; Kramer, R.M.; Barnes, V.L.; Dowling, Q.M.; Vedvick, T.S. Working together: Interactions between vaccine antigens and adjuvants. Ther. Adv. Vaccine. 2013, 1, 7–20. [Google Scholar] [CrossRef]
- McKee, A.S.; Munks, M.W.; Marrack, P. How do adjuvants work? Important considerations for new generation adjuvants. Immunity 2007, 27, 687–690. [Google Scholar] [CrossRef]
- Arrecubieta, C.; Lopez, R.; Garcia, E. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: Sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J. Bacteriol. 1994, 176, 6375–6383. [Google Scholar] [CrossRef]
- Hathaway, L.J.; Stutzmann Meier, P.; Battig, P.; Aebi, S.; Muhlemann, K. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J. Bacteriol. 2004, 186, 3721–3729. [Google Scholar] [CrossRef]
- Battig, P.; Muhlemann, K. Capsule genes of Streptococcus pneumoniae influence growth in vitro. FEMS Immunol. Med. Microbiol. 2007, 50, 324–329. [Google Scholar] [CrossRef]
- Iyer, R.; Baliga, N.S.; Camilli, A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 2005, 187, 8340–8349. [Google Scholar] [CrossRef]
- Hamaguchi, S.; Zafar, M.A.; Cammer, M.; Weiser, J.N. Capsule prolongs survival of Streptococcus pneumoniae during starvation. Infect. Immun. 2018. [Google Scholar] [CrossRef]
- Sanchez, C.J.; Kumar, N.; Lizcano, A.; Shivshankar, P.; Dunning Hotopp, J.C.; Jorgensen, J.H.; Tettelin, H.; Orihuela, C.J. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS ONE 2011, 6, e28738. [Google Scholar] [CrossRef]
- Rubins, J.B.; Charboneau, D.; Paton, J.C.; Mitchell, T.J.; Andrew, P.W.; Janoff, E.N. Dual function of pneumolysin in the early pathogenesis of murine pneumococcal pneumonia. J. Clin. Invest. 1995, 95, 142–150. [Google Scholar] [CrossRef]
- Orihuela, C.J.; Gao, G.; Francis, K.P.; Yu, J.; Tuomanen, E.I. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 2004, 190, 1661–1669. [Google Scholar] [CrossRef]
- Briles, D.E.; Crain, M.J.; Gray, B.M.; Forman, C.; Yother, J. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 1992, 60, 111–116. [Google Scholar] [CrossRef]
- Marks, L.R.; Parameswaran, G.I.; Hakansson, A.P. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect. Immun. 2012, 80, 2744–2760. [Google Scholar] [CrossRef]
- Cherny, K.E.; Sauer, K. Pseudomonas aeruginosa requires the DNA-specific endonuclease EndA to degrade eDNA to disperse from the biofilm. J. Bacteriol. 2019. [Google Scholar] [CrossRef]
- Magee, A.D.; Yother, J. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 2001, 69, 3755–3761. [Google Scholar] [CrossRef]
- Hathaway, L.J.; Grandgirard, D.; Valente, L.G.; Tauber, M.G.; Leib, S.L. Streptococcus pneumoniae capsule determines disease severity in experimental pneumococcal meningitis. Open. Biol. 2016, 6. [Google Scholar] [CrossRef]
- Hyams, C.; Camberlein, E.; Cohen, J.M.; Bax, K.; Brown, J.S. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 2010, 78, 704–715. [Google Scholar] [CrossRef]
- Nelson, A.L.; Roche, A.M.; Gould, J.M.; Chim, K.; Ratner, A.J.; Weiser, J.N. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 2007, 75, 83–90. [Google Scholar] [CrossRef]
- Krone, C.L.; Trzcinski, K.; Zborowski, T.; Sanders, E.A.; Bogaert, D. Impaired innate mucosal immunity in aged mice permits prolonged Streptococcus pneumoniae colonization. Infect. Immun. 2013, 81, 4615–4625. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- von Kockritz-Blickwede, M.; Goldmann, O.; Thulin, P.; Heinemann, K.; Norrby-Teglund, A.; Rohde, M.; Medina, E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008, 111, 3070–3080. [Google Scholar] [CrossRef]
- Ochsenbein, A.F.; Fehr, T.; Lutz, C.; Suter, M.; Brombacher, F.; Hengartner, H.; Zinkernagel, R.M. Control of early viral and bacterial distribution and disease by natural antibodies. Science 1999, 286, 2156–2159. [Google Scholar] [CrossRef]
- Murakami, A.; Moriyama, H.; Osako-Kabasawa, M.; Endo, K.; Nishimura, M.; Udaka, K.; Muramatsu, M.; Honjo, T.; Azuma, T.; Shimizu, T. Low-affinity IgM antibodies lacking somatic hypermutations are produced in the secondary response of C57BL/6 mice to (4-hydroxy-3-nitrophenyl)acetyl hapten. Int. Immunol. 2014, 26, 195–208. [Google Scholar] [CrossRef]
- Leliefeld, P.H.; Koenderman, L.; Pillay, J. How Neutrophils Shape Adaptive Immune Responses. Front. Immunol. 2015, 6, 471. [Google Scholar] [CrossRef]
- Ramos-Sevillano, E.; Ercoli, G.; Brown, J.S. Mechanisms of Naturally Acquired Immunity to Streptococcus pneumoniae. Front. Immunol. 2019, 10, 358. [Google Scholar] [CrossRef]
- Prevaes, S.M.; van Wamel, W.J.; de Vogel, C.P.; Veenhoven, R.H.; van Gils, E.J.; van Belkum, A.; Sanders, E.A.; Bogaert, D. Nasopharyngeal colonization elicits antibody responses to staphylococcal and pneumococcal proteins that are not associated with a reduced risk of subsequent carriage. Infect. Immun. 2012, 80, 2186–2193. [Google Scholar] [CrossRef]
- Wilson, R.; Cohen, J.M.; Reglinski, M.; Jose, R.J.; Chan, W.Y.; Marshall, H.; de Vogel, C.; Gordon, S.; Goldblatt, D.; Petersen, F.C.; et al. Naturally Acquired Human Immunity to Pneumococcus Is Dependent on Antibody to Protein Antigens. PLoS Pathog. 2017, 13, e1006137. [Google Scholar] [CrossRef]
- Turner, P.; Turner, C.; Green, N.; Ashton, L.; Lwe, E.; Jankhot, A.; Day, N.P.; White, N.J.; Nosten, F.; Goldblatt, D. Serum antibody responses to pneumococcal colonization in the first 2 years of life: Results from an SE Asian longitudinal cohort study. Clin. Microbiol. Infect. 2013, 19, E551–E558. [Google Scholar] [CrossRef]
- Martinot, M.; Oswald, L.; Parisi, E.; Etienne, E.; Argy, N.; Grawey, I.; De Briel, D.; Zadeh, M.M.; Federici, L.; Blaison, G.; et al. Immunoglobulin deficiency in patients with Streptococcus pneumoniae or Haemophilus influenzae invasive infections. Int. J. Infect. Dis. 2014, 19, 79–84. [Google Scholar] [CrossRef]
- Klein, C.; Lisowska-Grospierre, B.; LeDeist, F.; Fischer, A.; Griscelli, C. Major histocompatibility complex class II deficiency: Clinical manifestations, immunologic features, and outcome. J. Pediatr. 1993, 123, 921–928. [Google Scholar] [CrossRef]
- Briles, D.E.; Miyaji, E.; Fukuyama, Y.; Ferreira, D.M.; Fujihashi, K. Elicitation of mucosal immunity by proteins of Streptococcus pneumoniae. Adv. Otorhinolaryngol. 2011, 72, 25–27. [Google Scholar] [CrossRef]
- Ibrahim, Y.M. Attenuated HtrA-mutant of Streptococcus pneumoniae induces protection in murine models of pneumococcal pneumonia and bacteraemia. Afr. J. Microbiol. Res. 2013, 7, 237–244. [Google Scholar]
- Ibrahim, Y.M.; Kerr, A.R.; McCluskey, J.; Mitchell, T.J. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 2004, 72, 3584–3591. [Google Scholar] [CrossRef]
- Cohen, J.M.; Wilson, R.; Shah, P.; Baxendale, H.E.; Brown, J.S. Lack of cross-protection against invasive pneumonia caused by heterologous strains following murine Streptococcus pneumoniae nasopharyngeal colonisation despite whole cell ELISAs showing significant cross-reactive IgG. Vaccine 2013, 31, 2328–2332. [Google Scholar] [CrossRef]
- Kim, J.O.; Romero-Steiner, S.; Sørensen, U.B.S.; Blom, J.; Carvalho, M.; Barnard, S.; Carlone, G.; Weiser, J.N. Relationship between Cell Surface Carbohydrates and Intrastrain Variation on Opsonophagocytosis ofStreptococcus pneumoniae. Infect. Immun. 1999, 67, 2327–2333. [Google Scholar] [CrossRef]
- Jeong, D.G.; Jeong, E.S.; Seo, J.H.; Heo, S.H.; Choi, Y.K. Difference in Resistance to Streptococcus pneumoniae Infection in Mice. Lab. Anim. Res. 2011, 27, 91–98. [Google Scholar] [CrossRef]
- Gingles, N.A.; Alexander, J.E.; Kadioglu, A.; Andrew, P.W.; Kerr, A.; Mitchell, T.J.; Hopes, E.; Denny, P.; Brown, S.; Jones, H.B.; et al. Role of genetic resistance in invasive pneumococcal infection: Identification and study of susceptibility and resistance in inbred mouse strains. Infect. Immun. 2001, 69, 426–434. [Google Scholar] [CrossRef]
- Kadioglu, A.; Andrew, P.W. The innate immune response to pneumococcal lung infection: The untold story. Trends Immunol. 2004, 25, 143–149. [Google Scholar] [CrossRef]
- Storisteanu, D.M.; Pocock, J.M.; Cowburn, A.S.; Juss, J.K.; Nadesalingam, A.; Nizet, V.; Chilvers, E.R. Evasion of Neutrophil Extracellular Traps by Respiratory Pathogens. Am. J. Respir. Cell Mol. Biol. 2017, 56, 423–431. [Google Scholar] [CrossRef]





| Strains | LD50 (CFU) | Fold of Attenuation * |
|---|---|---|
| SPEC | 3.5 × 1010 | 23 |
| Wild-type D39 | 1.5 × 109 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amonov, M.; Simbak, N.; Wan Hassan, W.M.R.; Ismail, S.; A. Rahman, N.I.; Clarke, S.C.; Yeo, C.C. Disruption of the cpsE and endA Genes Attenuates Streptococcus pneumoniae Virulence: Towards the Development of a Live Attenuated Vaccine Candidate. Vaccines 2020, 8, 187. https://doi.org/10.3390/vaccines8020187
Amonov M, Simbak N, Wan Hassan WMR, Ismail S, A. Rahman NI, Clarke SC, Yeo CC. Disruption of the cpsE and endA Genes Attenuates Streptococcus pneumoniae Virulence: Towards the Development of a Live Attenuated Vaccine Candidate. Vaccines. 2020; 8(2):187. https://doi.org/10.3390/vaccines8020187
Chicago/Turabian StyleAmonov, Malik, Nordin Simbak, Wan Mohd. Razin Wan Hassan, Salwani Ismail, Nor Iza A. Rahman, Stuart C. Clarke, and Chew Chieng Yeo. 2020. "Disruption of the cpsE and endA Genes Attenuates Streptococcus pneumoniae Virulence: Towards the Development of a Live Attenuated Vaccine Candidate" Vaccines 8, no. 2: 187. https://doi.org/10.3390/vaccines8020187
APA StyleAmonov, M., Simbak, N., Wan Hassan, W. M. R., Ismail, S., A. Rahman, N. I., Clarke, S. C., & Yeo, C. C. (2020). Disruption of the cpsE and endA Genes Attenuates Streptococcus pneumoniae Virulence: Towards the Development of a Live Attenuated Vaccine Candidate. Vaccines, 8(2), 187. https://doi.org/10.3390/vaccines8020187






