Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Infection of Cells
2.3. Flow Cytometry Analysis of Infected Cells
2.4. Isolation of MHC I-Bound Peptides and Mass Spectrometric Analysis
2.5. Database Search and Filtering
2.6. Peptide Synthesis
2.7. Immunization of Mice
2.8. IFN-γ ELISPOT Assay
2.9. MHC Dextramer Staining
2.10. Intracellular Cytokine Staining (ICS)
2.11. In Vitro Re-Stimulation of Splenocytes
2.12. Statistical Analysis
3. Results
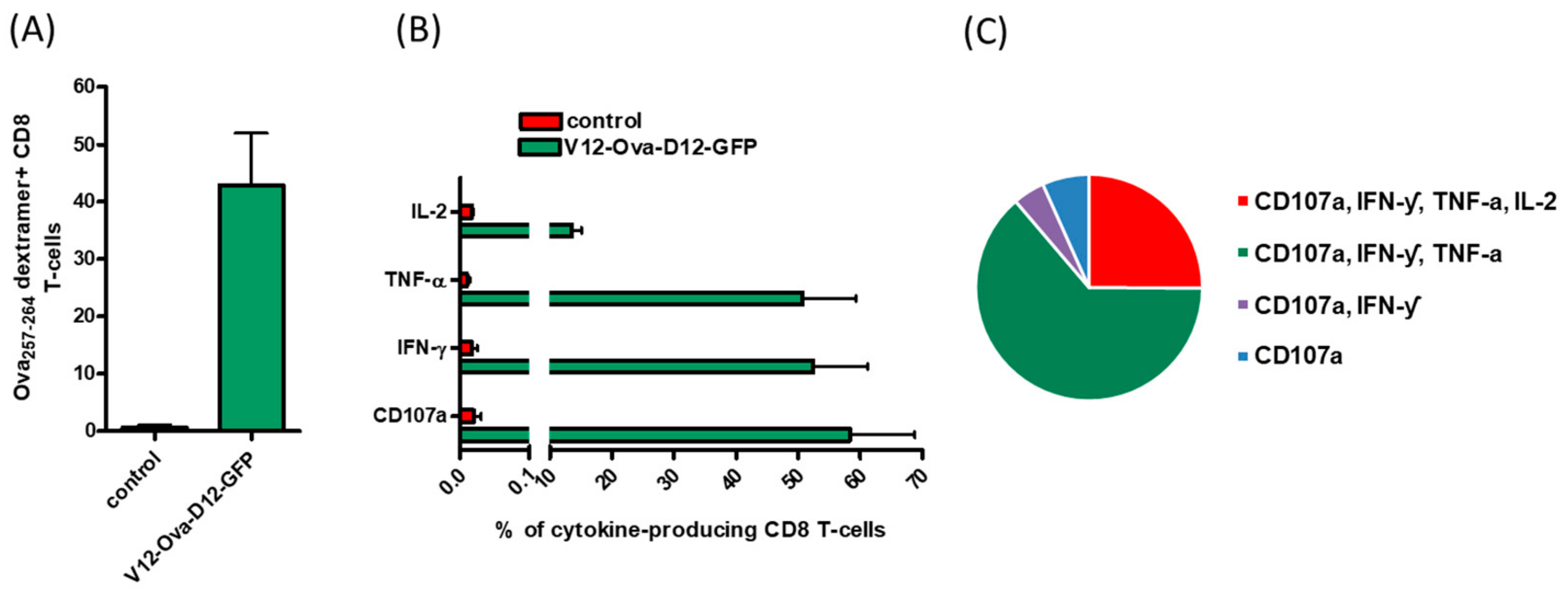
3.1. ORFV Vector Strain D1701-V Efficiently Induces Transgene-Specific CD8+ T Cell Response
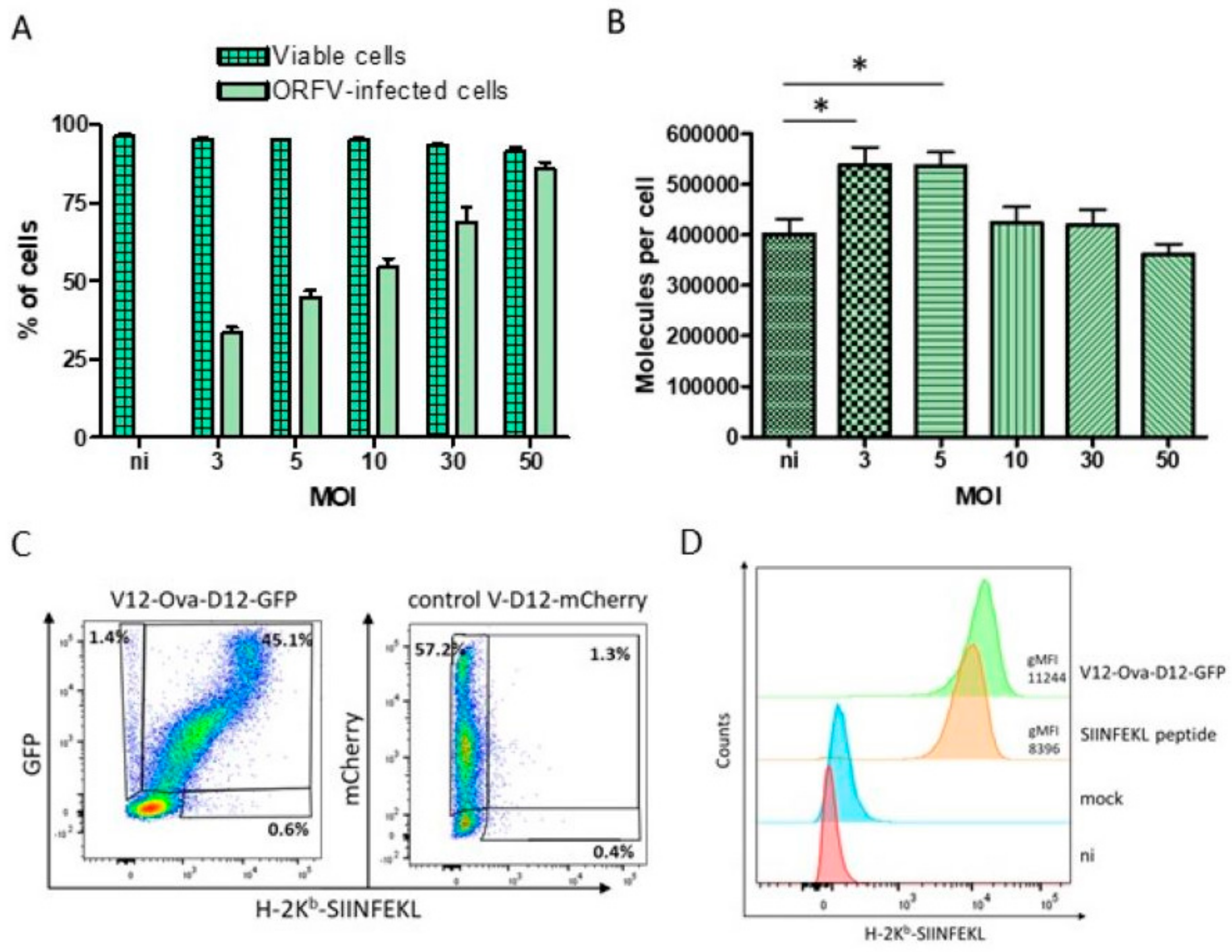
3.2. HeLa-Kb Cells Are Suitable Targets for the Identification of ORFV-Derived Peptides
3.3. Identification of ORFV-Derived H-2Kb-Presented Peptides by LC-MS/MS
3.4. Characteristics of ORFV-Derived H-2Kb-Presented Peptides
3.5. Characteristics of ORFV-Derived H-2Kb-Presented Peptides
3.6. ICS Validates Absence of CD8+ T Cell Responses to the Individual ORFV-Derived Peptides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Folgori, A.; Capone, S. Engineered viruses as vaccine platforms. Innov. Vaccinol. 2012, 65–86. [Google Scholar] [CrossRef]
- Nayak, S.; Herzog, R.W. Progress and prospects: Immune responses to viral vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gordo, E.; Podgorski, I.I.; Downes, N.; Alemany, R. Circumventing antivector immunity: Potential use of nonhuman adenoviral vectors. Hum. Gene Ther. 2014, 25, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.M.; Mercer, A.A. Ovine diseases. Orf. Vet Res. 1998, 29, 311–326. [Google Scholar] [PubMed]
- Büttner, M.; Rziha, H.J. Parapoxviruses: From the lesion to the viral genome. J. Vet. Med. B Infect. Dis. Vet. Public Health 2002, 49, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hussain, K.A.; Burger, D. In vivo and in vitro characteristics of contagious ecthyma virus isolates: Host response mechanism. Vet. Microbiol. 1989, 19, 23–36. [Google Scholar] [CrossRef]
- Haig, D.M. Orf virus infection and host immunity. Curr. Opin. Infect. Dis. 2006, 19, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Fachinger, V.; Schlapp, T.; Strube, W.; Schmeer, N.; Saalmuller, A. Poxvirus-induced immunostimulating effects on porcine leukocytes. J. Virol. 2000, 74, 7943–7951. [Google Scholar] [CrossRef] [PubMed]
- Friebe, A.; Siegling, A.; Friederichs, S.; Volk, H.D.; Weber, O. Immunomodulatory effects of inactivated parapoxvirus ovis (ORF virus) on human peripheral immune cells: Induction of cytokine secretion in monocytes and Th1-like cells. J. Virol. 2004, 78, 9400–9411. [Google Scholar] [CrossRef] [PubMed]
- Rziha, H.J.; Henkel, M.; Cottone, R.; Bauer, B.; Auge, U.; Götz, F.; Pfaff, E.; Röttgen, M.; Dehio, C.; Büttner, M. Generation of recombinant parapoxviruses: Non-essential genes suitable for insertion and expression of foreign genes. J. Biotechnol. 2000, 83, 137–145. [Google Scholar] [CrossRef]
- Rziha, H.J.; Büttner, M.; Müller, M.; Salomon, F.; Reguzova, A.; Laible, D.; Amann, R. Genomic characterization of Orf virus strain D1701-V (parapoxvirus) and development of novel sites for multiple transgene expression. Viruses 2019, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.; Planz, O.; Stitz, L.; Rziha, H.J. Novel recombinant parapoxvirus vectors induce protective humoral and cellular immunity against lethal herpesvirus challenge infection in mice. J. Virol. 2003, 77, 9312–9323. [Google Scholar] [CrossRef] [PubMed]
- Rohde, J.; Schirrmeier, H.; Granzow, H.; Rziha, H.J. A new recombinant Orf virus (ORFV, parapoxvirus) protects rabbits against lethal infection with rabbit hemorrhagic disease virus (RHDV). Vaccine 2011, 29, 9256–9264. [Google Scholar] [CrossRef] [PubMed]
- Voigt, H.; Merant, C.; Wienhold, D.; Braun, A.; Hutet, E.; Le Potier, M.F.; Saalmuller, A.; Pfaff, E.; Büttner, M. Efficient priming against classical swine fever with a safe glycoprotein E2 expressing Orf virus recombinant (ORFV VrV-E2). Vaccine 2007, 25, 5915–5926. [Google Scholar] [CrossRef] [PubMed]
- Henkel, M.; Planz, O.; Fischer, T.; Stitz, L.; Rziha, H.J. Prevention of virus persistence and protection against immunopathology after Borna disease virus infection of the brain by a novel Orf virus recombinant. J. Virol. 2005, 79, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Dory, D.; Fischer, T.; Beven, V.; Cariolet, R.; Rziha, H.J.; Jestin, A. Prime-boost immunization using DNA vaccine and recombinant Orf virus protects pigs against pseudorabies virus (herpes suid 1). Vaccine 2006, 24, 6256–6263. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.M.; Rijsewijk, F.A.; Moonen-Leusen, H.W.; Bianchi, A.T.; Rziha, H.J. Comparison of different prime-boost regimes with DNA and recombinant Orf virus based vaccines expressing glycoprotein D of pseudorabies virus in pigs. Vaccine 2010, 28, 1808–1813. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.; Rohde, J.; Wulle, U.; Conlee, D.; Raue, R.; Martinon, O.; Rziha, H.J. A new rabies vaccine based on a recombinant Orf virus (Parapoxvirus) expressing the rabies virus glycoprotein. J. Virol. 2013, 87, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Rohde, J.; Amann, R.; Rziha, H.J. New Orf virus (parapoxvirus) recombinant expressing H5 hemagglutinin protects mice against H5N1 and H1N1 influenza a virus. PLoS ONE 2013, 8, e83802. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Müller, M.; Yigitliler, A.; Xi, J.; Simon, C.; Feger, T.; Rziha, H.J.; Stubenrauch, F.; Rammensee, H.G.; Iftner, T.; et al. An Orf Virus-based therapeutic vaccine for the treatment of papillomavirus-induced tumors. J. Virol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.B.; Gill, H.S.; Haig, D.M.; Husband, A.J. In vivo T-cell subset depletion suggests that T-cells and a humoral immune response are important for the elimination of Orf virus from the skin of sheep. Vet. Immunol. Immunopathol. 2000, 74, 249–262. [Google Scholar] [CrossRef]
- Haig, D.M.; Entrican, G.; Yirrell, D.L.; Deane, D.; Millar, H.R.P.; Norval, M.; Reid, H.W. Differential appearance of interferon and colony stimulating activity in afferent versus efferent lymph following Orf virus infection in sheep. Vet. Dermatol. 1992, 3, 221–229. [Google Scholar] [CrossRef]
- Haig, D.M.; Deane, D.; Percival, A.; Myatt, N.; Thomson, J.; Inglis, L.; Rothel, J.; Heng-Fong, S.; Wood, P.; Millar, H.R.P.; et al. The cytokine response of afferent lymph following Orf virus reinfection of sheep. Vet. Dermatol. 1996, 7, 11–20. [Google Scholar] [CrossRef]
- Haig, D.M.; Hutchison, G.; Thomson, J.; Yirrell, D.; Reid, H.W. Cytolytic activity and associated serine protease expression by skin and afferent lymph CD8+ T cells during Orf virus reinfection. J. Gen. Virol. 1996, 77, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.M.; McInnes, C.J.; Hutchison, G.; Seow, H.F.; Reid, H.W. Cyclosporin A abrogates the acquired immunity to cutaneous reinfection with the parapoxvirus orf virus. Immunology 1996, 89, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.M.; Hopkins, J.; Miller, H.R.P. Local immune responses in afferent and efferent lymph. Immunology 1999, 96, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Buddle, B.M.; Dellers, R.W.; Schurig, G.G. Contagious ecthyma virus-vaccination failures. Am. J. Vet. Res. 1984, 45, 263–266. [Google Scholar] [PubMed]
- Yirrell, D.L.; Vestey, J.; Norval, M. Immune responses of patients to orf virus infection. Br. J. Dermatol. 1994, 130, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Schirmbeck, R.; Reimann, J.; Kochanek, S.; Kreppel, F. The immunogenicity of adenovirus vectors limits the multispecificity of CD8 T-cell responses to vector-encoded transgenic antigens. Mol. Ther. 2008, 16, 1609–1616. [Google Scholar] [CrossRef]
- Sumida, S.M.; Truitt, D.M.; Kishko, M.G.; Arthur, J.C.; Jackson, S.S.; Gorgone, D.A.; Lifton, M.A.; Koudstaal, W.; Pau, M.G.; Kostense, S.; et al. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J. Virol. 2004, 78, 2666–2673. [Google Scholar] [CrossRef] [PubMed]
- Altenburg, A.F.; van Trierum, S.E.; de Bruin, E.; de Meulder, D.; van de Sandt, C.E.; van der Klis, F.R.M.; Fouchier, R.A.M.; Koopmans, M.P.G.; Rimmelzwaan, G.F.; de Vries, R.D. Effects of pre-existing orthopoxvirus-specific immunity on the performance of modified vaccinia virus ankara-based influenza vaccines. Sci. Rep. 2018, 8, 6474. [Google Scholar] [CrossRef] [PubMed]
- McKelvey, T.; Tang, A.; Bett, A.J.; Casimiro, D.R.; Chastain, M. T-cell response to adenovirus hexon and DNA-binding protein in mice. Gene Ther. 2004, 11, 791–796. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roederer, M.; Koup, R.A. Optimized determination of T cell epitope responses. J. Immunol. Methods. 2003, 274, 221–228. [Google Scholar] [CrossRef]
- Fiore-Gartland, A.; Manso, B.A.; Friedrich, D.P.; Gabriel, E.E.; Finak, G.; Moodie, Z.; Hertz, T.; De Rosa, S.C.; Frahm, N.; Gilbert, P.B.; et al. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: An evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PLoS ONE 2016, 11, e0147812. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.S.; Berzofsky, J.A. From genome to vaccine-new immunoinformatics tools for vaccine design. Methods 2004, 34, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Moutaftsi, M.; Peters, B.; Pasquetto, V.; Tscharke, D.C.; Sidney, J.; Bui, H.H.; Grey, H.; Sette, A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006, 24, 817–819. [Google Scholar] [CrossRef] [PubMed]
- St Leger, A.J.; Peters, B.; Sidney, J.; Sette, A.; Hendricks, R.L. Defining the herpes simplex virus-specific CD8+ T cell repertoire in C57BL/6 mice. J. Immunol. 2011, 186, 3927–3933. [Google Scholar] [CrossRef] [PubMed]
- Wölk, B.; Trautwein, C.; Büchele, B.; Kersting, N.; Blum, H.E.; Rammensee, H.G.; Cerny, A.; Stevanovic, A.; Moradpour, D.; Brass, V. Identification of naturally processed hepatitis C virus-derived major histocompatibility complex class I ligands. PLoS ONE 2012, 7, e29286. [Google Scholar] [CrossRef] [PubMed]
- Rötzschke, O.; Falk, K.; Deres, K.; Schild, H.; Norda, M.; Metzger, J.; Jung, G.; Rammensee, H.G. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 1990, 348, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Ternette, N.; Block, P.D.; Sánchez-Bernabéu, Á.; Borthwick, N.; Pappalardo, E.; Abdul-Jawad, S.; Ondondo, B.; Charles, P.D.; Dorrell, L.; Kessler, B.M.; et al. Early kinetics of the HLA class I-Associated peptidome of MVA.HIVconsv-infected cells. J. Virol. 2015, 89, 5760–5771. [Google Scholar] [CrossRef] [PubMed]
- Lübke, M.; Spalt, S.; Kowalewski, D.J.; Zimmermann, C.; Bauersfeld, L.; Nelde, A.; Bichmann, L.; Marcu, A.; Peper, J.K.; Kohlbacher, O.; et al. Identification of HCMV-derived T cell epitopes in seropositive individuals through viral deletion models. J. Exp. Med. 2020, 217, e20191164. [Google Scholar] [CrossRef] [PubMed]
- Purcell, A.W.; Ramarathinam, S.H.; Ternette, N. Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 2019, 14, 1687–1707. [Google Scholar] [CrossRef] [PubMed]
- Croft, N.P.; Smith, S.A.; Pickering, J.; Sidney, J.; Peters, B.; Faridi, P.; Witney, M.J.; Sebastian, P.; Flesch, I.E.A.; Heading, S.L.; et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA 2019, 116, 3112–3117. [Google Scholar] [CrossRef] [PubMed]
- Gilchuk, P.; Spencer, C.T.; Conant, S.B.; Hill, T.; Gray, J.J.; Niu, X.; Zheng, M.; Erickson, J.J.; Boyd, K.L.; McAfee, K.J.; et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J. Clin. Invest. 2013, 123, 1976–1987. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.; Rötzschke, O.; Faath, S.; Goth, S.; Graef, I.; Shastri, N.; Rammensee, H.G. Both human and mouse cells expressing H-2Kb and ovalbumin process the same peptide, SIINFEKL. Cell Immunol. 1993, 150, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.; Rötzschke, O.; Stevanović, S.; Jung, G.; Rammensee, H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991, 351, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Kowalewski, D.J.; Stevanović, S. Purification and identification of naturally presented MHC Class I and II ligands. Methods Mol. Biol. 2019, 1988, 123–136. [Google Scholar] [PubMed]
- Delhon, G.; Tulman, E.R.; Afonso, C.L.; Lu, Z.; de la Concha-Bermejillo, A.; Lehmkuhl, H.D.; Piccone, M.E.; Kutish, G.F.; Rock, D.L. Genomes of the parapoxviruses Orf virus and bovine papular stomatitis virus. J. Virol. 2003, 78, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Di Marco, M.; Stevanović, S. Identification of MHC ligands and establishing MHC class I peptide motifs. Methods Mol. Biol. 2019, 1988, 137–147. [Google Scholar] [PubMed]
- McGuire, M.J.; Johnston, S.A.; Sykes, K.F. Novel immune-modulator identified by a rapid, functional screen of the parapoxvirus ovis (Orf virus) genome. Proteome Sci. 2012, 10, 4. [Google Scholar] [CrossRef]
- Broyles, S.S. Vaccinia virus transcription. J. Gen. Virol. 2003, 84, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Greenbaum, J.A.; Sundström, M.; Schaffer, L.; Hammond, J.A.; Pasquetto, V.; Oseroff, C.; Hendrickson, R.C.; Lefkowitz, E.J.; Tscharke, D.C.; et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. USA 2008, 105, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Nanda, A.; Havenga, M.J.; Abbink, P.; Lynch, D.M.; Ewald, B.A.; Liu, J.; Thorner, A.R.; Swanson, P.E.; Gorgone, D.A.; et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 2006, 441, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune responses to viral gene therapy vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Ura, T.; Okuda, K.; Shimada, M. Developments in viral vector-based vaccines. Vaccines 2014, 2, 624–641. [Google Scholar] [CrossRef]
- Sette, A.; Grey, H.; Oseroff, C.; Peters, B.; Moutaftsi, M.; Crotty, S.; Assarsson, E.; Greenbaum, J.; Kim, Y.; Kolla, R.; et al. Definition of epitopes and antigens recognized by vaccinia specific immune responses: Their conservation in variola virus sequences, and use as a model system to study complex pathogens. Vaccine 2009, 27, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.C.W.; Flesch, I.E.A.; Tscharke, D.C. Immunodomination during peripheral vaccinia virus infection. PLoS Pathog. 2013, 9, e1003329. [Google Scholar] [CrossRef] [PubMed]
- Flesch, I.E.A.; Hollett, N.A.; Wong, Y.C.; Quinan, B.R.; Howard, D.; da Fonseca, F.G.; Tscharke, D.C. Extent of systemic spread determines CD8+ T Cell immunodominance for laboratory strains, smallpox vaccines, and zoonotic isolates of vaccinia virus. J. Immunol. 2015, 195, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Croft, N.P.; Smith, S.A.; Wong, Y.C.; Tan, C.T.; Dudek, N.L.; Flesch, I.E.A.; Lin, L.C.W.; Tscharke, D.C.; Purcell, A.W. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 2013, 9, e1003129. [Google Scholar] [CrossRef] [PubMed]
- Brun, A.; Albina, E.; Barret, T.; Chapmanc, D.A.G.; Czub, M.; Dixon, L.K.; Keil, G.M.; Klonjkowski, B.; Le Potier, M.F.; Libeau, G.; et al. Antigen delivery systems for veterinary vaccine development. Viral-vector based delivery systems. Vaccine 2008, 26, 6508–6528. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.M. Subversion and piracy: DNA viruses and immune evasion. Res. Vet. Sci. 2001, 70, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Maruri-Avidal, L.; Weisberg, A.S.; Moss, B. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J. Virol. 2011, 85, 12431–12441. [Google Scholar] [CrossRef] [PubMed]


| No. | Sequence | Length (aa) | UniProt Accession No. | Antigen/Protein * | Position | NetMHC Binding Affinity (nM) | NetMHC % Rank of Predicted Affinity |
|---|---|---|---|---|---|---|---|
| 1 | AIYGFGVTF | 9 | ADY76886.1; ADY76885.1 | ORFV056 RNA-polymerase subunit RPO147 | 569–577 | 248.6 | 0.5757 |
| 2 | VGYPRQNAV | 9 | ADY76846.1; ADY76844.1 | ORFV097 DNA-polymerase processivity factor | 364–372 | 95.7 | 0.2477 |
| 3 | IGYMVKNL | 8 | ADY76734.1 | ORFV075 Rifampicin resistance protein | 212–219 | 19.1 | 0.0441 |
| 4 | VGFVHPIAM | 9 | ADY76731.1 | ORF079 Virion core protein P4b | 258–266 | 51.9 | 0.1255 |
| 5 | SILKFEERL | 9 | ADY76728.1 | ORF083 Early transcription factor VETFL | 286–294 | 93.8 | 0.2427 |
| 6 | LNLMYPNI | 8 | ADY76725.1 | ORFV86 Virion core precursor protein P4a | 172–179 | 26.7 | 0.0674 |
| 7 | SGSVPYARL | 9 | ADY76722.1 | ORFV090 IMV membrane protein | 73–81 | 140.6 | 0.3490 |
| 8 | AAFEFRDL | 8 | ADY76890.1 | ORF052 putative IMV membrane protein | 72–79 | 12.6 | 0.0253 |
| 9 | VGFVHPIA | 8 | ADY76731.1 | ORF079 Virion core protein P4b | 258–265 | 485.9 | 0.9853 |
| 10 | KILAPFNFL | 9 | ADY76886.1; ADY76885.1 | ORFV056 RNA-polymerase subunit RPO147 | 834–842 | 385.2 | 0.8315 |
| 11 | ISALFKQL | 8 | ADY76725.1; ADY76726.1 | ORFV086 Virion core protein P4a | 781–788 | 14.2 | 0.0296 |
| 12 | TEFPVFEEL | 9 | ADY76883.1 | ORFV058 (IMV, viral entry) | 182–190 | 3095.8 | 4.6680 |
| 13 | VNIVRQEEL | 9 | ADY76792.1 | ORFV016 Unknown | 35–43 | 270.1 | 0.6179 |
| 14 | AIIKYTDL | 8 | ADY76837.1 | ORFV110 EEV glycoprotein | 37–44 | 89.3 | 0.2306 |
| 15 | SILERYNLF | 9 | ADY76725.1; ADY76726.1 | ORFV086 Virion core protein P4a | 810–818 | 130.5 | 0.3285 |
| 16 | VFFRVTVL | 8 | ADY76779.1 | ORFV028 DNA-binding protein | 644–651 | 73.4 | 0.1860 |
| 17 | GSVPYARL | 8 | ADY76722.1 | ORFV090 IMV membrane protein | 74–81 | 55.2 | 0.1387 |
| 18 | QNYSYSERLL | 10 | ADY76809.1 | ORFV129 Ankyrin/F-box protein | 115–124 | 180.9 | 0.4297 |
| 19 | RVNTFTAV | 8 | ADY76793.1 | ORFV015 Unknown | 33–40 | 738.0 | 1.4130 |
| 20 | TAVDFTQFL | 9 | ADY76778.1 | ORFV029 Unknown | 244–252 | 172.0 | 0.4073 |
| 21 | QNYSYSERL | 9 | ADY76809.1 | ORFV129 Ankyrin/F-box protein | 115–123 | 24.3 | 0.0592 |
| 22 | AIYAFRLT | 8 | ADY76719.1 | ORFV094 Phosphorylated IMV membrane protein | 160–167 | 150.9 | 0.3685 |
| 23 | ANVDFMEYV | 9 | ADY76886.1; ADY76885.1 | ORFV056 RNA-polymerase subunit RPO147 | 880–888 | 397.0 | 0.8488 |
| 24 | ISVMFNNV | 8 | ADY76728.1 | ORF083 Early transcription factor VETFL | 484–491 | 10.5 | 0.0198 |
| 25 | VIFGRQPSL | 9 | ADY76886.1; ADY76885.1 | ORFV056 RNA-polymerase subunit RPO147 | 374–382 | 59.9 | 0.1538 |
| 26 | EQFSFSNV | 8 | ADY76841.1 | ORFV101 RNA-polymerase subunit RPO132 | 623–630 | 259.0 | 0.5952 |
| 27 | LIREFANL | 8 | ADY76728.1 | ORF083 Early transcription factor VETFL | 294–301 | 19.5 | 0.0456 |
| 28 | IAPQLRSL | 8 | ADY76724.1 | ORF088 Virion core protein | 24–31 | 418.0 | 0.8785 |
| 29 | ISIPRSVGF | 9 | ADY76841.1 | ORFV101 RNA-polymerase subunit RPO132 | 427–435 | 195.8 | 0.4646 |
| 30 | SIAPMNTGF | 9 | ADY76778.1 | ORFV029 Unknown | 216–224 | 3273.7 | 4.8979 |
| 31 | IAPQLRSLL | 9 | ADY76724.1 | ORF088 Virion core protein | 24–32 | 331.2 | 0.7371 |
| 32 | SATQFQSV | 8 | ADY76731.1 | ORF079 Virion core protein P4b | 230–237 | 482.6 | 0.9800 |
| 33 | SFVVVAQI | 8 | ADY76769.1 | ORFV040 Glutaredoxin-like protein | 98–105 | 2254.5 | 3.5474 |
| 34 | SIVSFKPTL | 9 | ADY76731.1 | ORF079 Virion core protein P4b | 386–394 | 75.0 | 0.1902 |
| 35 | TNVEIGKL | 8 | ADY76729.1 | ORFV082 Unknown | 277–284 | 1825.4 | 2.9640 |
| 36 | VIEIFKQL | 8 | ADY76732.1 | ORFV077 Late transcription factor VLTF-3 | 130–137 | 216.0 | 0.5087 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reguzova, A.; Ghosh, M.; Müller, M.; Rziha, H.-J.; Amann, R. Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes. Vaccines 2020, 8, 295. https://doi.org/10.3390/vaccines8020295
Reguzova A, Ghosh M, Müller M, Rziha H-J, Amann R. Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes. Vaccines. 2020; 8(2):295. https://doi.org/10.3390/vaccines8020295
Chicago/Turabian StyleReguzova, Alena, Michael Ghosh, Melanie Müller, Hanns-Joachim Rziha, and Ralf Amann. 2020. "Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes" Vaccines 8, no. 2: 295. https://doi.org/10.3390/vaccines8020295
APA StyleReguzova, A., Ghosh, M., Müller, M., Rziha, H.-J., & Amann, R. (2020). Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes. Vaccines, 8(2), 295. https://doi.org/10.3390/vaccines8020295





