Investigating Virological, Immunological, and Pathological Avenues to Identify Potential Targets for Developing COVID-19 Treatment and Prevention Strategies
Abstract
1. Introduction
2. Coronaviruses, Origin and Genome Structure
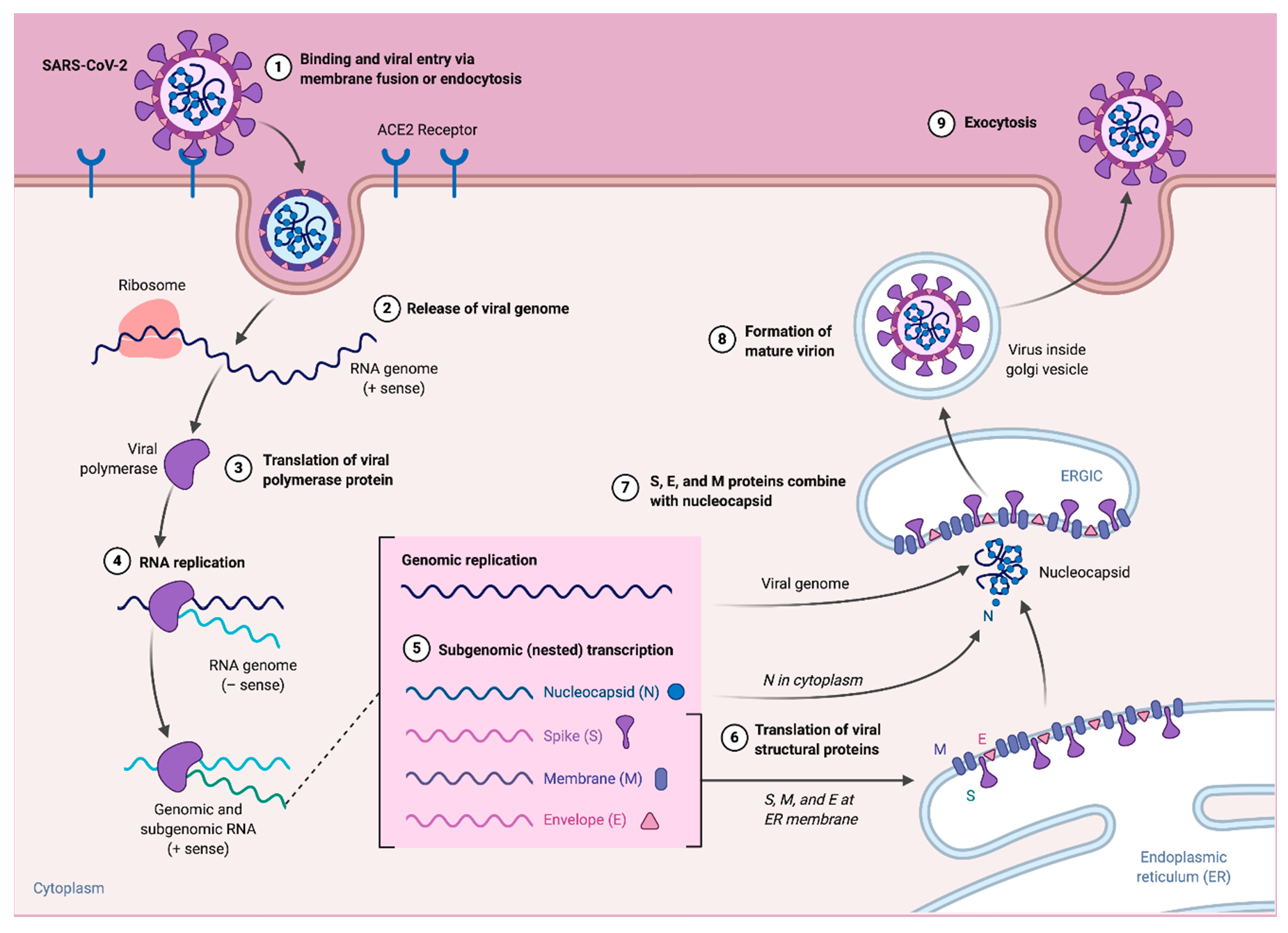
3. Replication and Pathogenesis
4. Clinical Characterization
5. Innate Immunity during COVID-19
6. Adaptive Immunity during COVID-19
6.1. T Cell Immune Response to SARS-CoV-2 Infection
6.2. Role of Memory T Cell against SARS-CoV-2
6.3. B Cell Immunity during SARS-CoV-2 Infection
6.4. B Cell Responses
7. Therapeutics
8. Vaccine Development
9. Antiviral Agents against Coronaviruses
10. Immune Cells and Therapeutic Perspectives
11. Convalescent Plasma Therapy for COVID-19
12. Neutralizing Monoclonal Antibodies
13. Cellular Therapy
14. CRISPR/Cas Mediated Therapeutic Options of COVID-19
15. Designer B and T Cells for Antiviral Activity
16. Diagnostic and Antiviral Potential of CRISPR/Cas13
17. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Frieman, M.B. Coronaviruses: Important emerging human pathogens. J. Virol. 2014, 88, 5209–5212. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- NBIC+. An Overview of Nanotechnology Patents Focusing on Coronaviruses. Available online: https://statnano.com/news/67513/An-Overview-of-Nanotechnology-Patents-Focusing-on-Coronaviruses (accessed on 10 March 2020).
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- WHO. WHO Announces COVID-19 Outbreak a Pandemic. Available online: http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic (accessed on 11 March 2020).
- World Health Organization. Coronavirus Disease (COVID-19) Situation Report—188. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200726-covid-19-sitrep-188.pdf?sfvrsn=f177c3fa_2 (accessed on 26 July 2020).
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020, 7. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Yin, Y.; Wunderink, R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018, 23, 130–137. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, evaluation and treatment coronavirus (COVID-19). In Statpearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Ahn, D.-G.; Shin, H.-J.; Kim, M.-H.; Lee, S.; Kim, H.-S.; Myoung, J.; Kim, B.-T.; Kim, S.-J. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef]
- Łoczechin, A.; Séron, K.; Barras, A.; Giovanelli, E.; Belouzard, S.; Chen, Y.-T.; Metzler-Nolte, N.; Boukherroub, R.; Dubuisson, J.; Szunerits, S. Functional Carbon Quantum Dots as Medical Countermeasures to Human Coronavirus. ACS Appl. Mater. Interfaces 2019, 11, 42964–42974. [Google Scholar] [CrossRef]
- Library, D.o.C.-P.H.I. Centers for Disease Control and Prevention. Available online: https://phil.cdc.gov/Details.aspx?pid=23354 (accessed on 28 April 2020).
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Abd Ellah, N.H.; Gad, S.F.; Muhammad, K.; E Batiha, G.; Hetta, H.F. Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against covid-19. Nanomedicine 2020. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Coronaviruses; Maier, H., Bickerton, E., Britton, P., Eds.; Methods in Molecular Biology: New York, NY, USA, 2015; Volume 1282, pp. 1–23. [Google Scholar]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.A.; Verkleij, A.; Rottier, P.J.M.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef]
- Neuman, B.W.; Adair, B.D.; Yoshioka, C.; Quispe, J.D.; Orca, G.; Kuhn, P.; Milligan, R.A.; Yeager, M.; Buchmeier, M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006, 80, 7918–7928. [Google Scholar] [CrossRef]
- Huang, X.; Li, M.; Xu, Y.; Zhang, J.; Meng, X.; An, X.; Sun, L.; Guo, L.; Shan, X.; Ge, J.; et al. Novel Gold Nanorod-Based HR1 Peptide Inhibitor for Middle East Respiratory Syndrome Coronavirus. ACS Appl. Mater. Interfaces 2019, 11, 19799–19807. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef]
- Yu, F.; Du, L.; Ojcius, D.M.; Pan, C.; Jiang, S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020, 22, 74–79. [Google Scholar] [CrossRef]
- Sawicki, S.; Sawicki, D. Coronavirus transcription: A perspective. In Coronavirus Replication and Reverse Genetics; Springer: Berlin, Germany, 2005; pp. 31–55. [Google Scholar]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020, 11, 216. [Google Scholar] [CrossRef]
- McGonagle, D.; O’Donnell, J.S.; Sharif, K.; Emery, P.; Bridgewood, C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020, 2, e437–e445. [Google Scholar] [CrossRef]
- Connors, J.M.; Levy, J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood 2020, 135, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Galbadage, T.; Peterson, B.M.; Gunasekera, R.S. Does COVID-19 Spread Through Droplets Alone? Front. Public Health 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Y.A.-O.; Zhang, A.L.; Wang, Y.A.-O.; Molina, M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 11857–14863. [Google Scholar] [CrossRef] [PubMed]
- Setti, L.; Passarini, F.; De Gennaro, G.; Barbieri, P.; Perrone, M.G.; Borelli, M.; Palmisani, J.; Di Gilio, A.; Piscitelli, P.; Miani, A. Airborne Transmission Route of COVID-19: Why 2 Meters/6 Feet of Inter-Personal Distance Could Not Be Enough. Int. J. Environ. Res. Public Health 2020, 17, 2932. [Google Scholar] [CrossRef]
- Harvala, H.; Robb, M.; Watkins, N.; Ijaz, S.; Dicks, S.; Patel, M.; Supasa, P.; Dejnirattisai, W.; Liu, C.; Mongkolsapaya, J.; et al. Convalescent plasma therapy for the treatment of patients with COVID-19: Assessment of methods available for antibody detection and their correlation with neutralising antibody levels. MedRxiv 2020. [Google Scholar] [CrossRef]
- Oberfeld, B.; Achanta, A.; Carpenter, K.; Chen, P.; Gilette, N.M.; Langat, P.; Said, J.T.; Schiff, A.E.; Zhou, A.S.; Barczak, A.K.; et al. SnapShot: COVID-19. Cell 2020, 181, 954. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Atluri, S.; Manchikanti, L.; Hirsch, J.A. Expanded Umbilical Cord Mesenchymal Stem Cells (UC-MSCs) as a Therapeutic Strategy in Managing Critically Ill COVID-19 Patients: The Case for Compassionate Use. Pain Physician 2020, 23, E71–E83. [Google Scholar] [PubMed]
- Zhao, X.; Zhang, B.; Li, P.; Ma, C.; Gu, J.; Hou, P.; Guo, Z.; Wu, H.; Bai, Y. Incidence, clinical characteristics and prognostic factor of patients with COVID-19: A systematic review and meta-analysis. MedRxiv 2020. [Google Scholar] [CrossRef]
- Qu, R.; Ling, Y.; Zhang, Y.H.; Wei, L.Y.; Chen, X.; Li, X.M.; Liu, X.Y.; Liu, H.M.; Guo, Z.; Ren, H.; et al. Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, W.; Guo, Y.; Chen, L.; Zhang, L.; Zhao, S.; Long, D.; Yu, L. Association between platelet parameters and mortality in coronavirus disease 2019: Retrospective cohort study. Platelets 2020, 31, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+ CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv 2020. [Google Scholar] [CrossRef]
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 2020, 6, 31. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-l.; Hui, D.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Zahran, A.M.; Zahran, Z.A.M.; El-Badawy, O.; Abdel-Rahim, M.H.; Ali, W.A.; Rayan, A.; El-Masry, M.A.; Abozaid, M.A.; Hetta, H.F. Prognostic impact of toll-like receptors 2 and 4 expression on monocytes in Egyptian patients with hepatocellular carcinoma. Immunol. Res. 2019, 67, 157–165. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- McKechnie, J.L.; Blish, C.A. The Innate Immune System: Fighting on the Front Lines or Fanning the Flames of COVID-19? Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fan, Y.; Lai, Y.; Han, T.; Li, Z.; Zhou, P.; Pan, P.; Wang, W.; Hu, D.; Liu, X.; et al. Coronavirus infections and immune responses. J. Med. Virol. 2020, 92, 424–432. [Google Scholar] [CrossRef]
- Hetta, H.F.; Mekky, M.A.; Zahran, A.M.; Abdel-Malek, M.O.; Ramadan, H.K.; Shafik, E.A.; Abbas, W.A.; Abbas El-Masry, M.; Mohamed, N.A.; Kamel, A.A.; et al. Regulatory B Cells and Their Cytokine Profile in HCV-Related Hepatocellular Carcinoma: Association with Regulatory T Cells and Disease Progression. Vaccines 2020, 8, 380. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Dassler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Pison, U.; Brand, M.; Joka, T.; Obertacke, U.; Bruch, J. Distribution and function of alveolar cells in multiply injured patients with trauma-induced ARDS. Intensive Care Med. 1988, 14, 602–609. [Google Scholar] [CrossRef]
- Park, W.B.; Kwon, N.J.; Choi, S.J.; Kang, C.K.; Choe, P.G.; Kim, J.Y.; Yun, J.; Lee, G.W.; Seong, M.W.; Kim, N.J.; et al. Virus Isolation from the First Patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020, 35, e84. [Google Scholar] [CrossRef]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 20, 269–270. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Yaqinuddin, A.; Kashir, J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med. Hypotheses 2020, 140, 109777. [Google Scholar] [CrossRef] [PubMed]
- Cecere, T.E.; Todd, S.M.; Leroith, T. Regulatory T cells in arterivirus and coronavirus infections: Do they protect against disease or enhance it? Viruses 2012, 4, 833–846. [Google Scholar] [CrossRef]
- Zahran, A.M.; Nafady-Hego, H.; Mansor, S.G.; Abbas, W.A.; Abdel-Malek, M.O.; Mekky, M.A.; Hetta, H.F. Increased frequency and FOXP3 expression of human CD8+ CD25High+ T lymphocytes and its relation to CD4 regulatory T cells in patients with hepatocellular carcinoma. Hum. Immunol. 2019, 80, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Zahran, A.M.; Mansor, S.G.; Abdel-Malek, M.O.; Mekky, M.A.; Abbas, W.A. Frequency and Implications of myeloid-derived suppressor cells and lymphocyte subsets in Egyptian patients with hepatitis C virus-related hepatocellular carcinoma. J. Med. Virol. 2019, 91, 1319–1328. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Chen, J.; Lau, Y.F.; Lamirande, E.W.; Paddock, C.D.; Bartlett, J.H.; Zaki, S.R.; Subbarao, K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010, 84, 1289–1301. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, J.; Perlman, S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010, 84, 9318–9325. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020. [Google Scholar] [CrossRef]
- Gimenez, E.; Albert, E.; Torres, I.; Remigia, M.J.; Alcaraz, M.J.; Galindo, M.J.; Blasco, M.L.; Solano, C.; Forner, M.J.; Redon, J.; et al. SARS-CoV-2-reactive interferon-gamma-producing CD8+ T cells in patients hospitalized with coronavirus disease 2019. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Mwafey, I.M.; Batiha, G.E.-S.; Alomar, S.Y.; Mohamed, N.A.; Ibrahim, M.A.; Elkady, A.; Meshaal, A.K.; Alrefai, H.; Khodeer, D.M.; et al. Cd19+ cd24hi cd38hi regulatory b cells and memory b cells in periodontitis: Association with pro-inflammatory and anti-inflammatory cytokines. Vaccines 2020, 8, 340. [Google Scholar] [CrossRef] [PubMed]
- Palm, A.-K.E.; Henry, C. Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Woodruff, M.; Ramonell, R.; Cashman, K.; Nguyen, D.; Ley, A.; Kyu, S.; Saini, A.; Haddad, N.; Chen, W.; Howell, J.C.; et al. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. MedRxiv 2020. [Google Scholar] [CrossRef]
- Jenks, S.A.; Cashman, K.S.; Zumaquero, E.; Marigorta, U.M.; Patel, A.V.; Wang, X.; Tomar, D.; Woodruff, M.C.; Simon, Z.; Bugrovsky, R.; et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 2018, 49, 725–739. [Google Scholar] [CrossRef]
- Mahmood, Z.; Muhammad, K.; Schmalzing, M.; Roll, P.; Dörner, T.; Tony, H.-P. Cd27-igd- memory b cells are modulated by in vivo interleukin-6 receptor (il-6r) blockade in rheumatoid arthritis. Arthritis Res. Ther. 2015, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.; Rattigan, S.M.; Borgert, B.; Moreno, C.; Solomon, B.D.; Rodriguez-Barraquer, I.; et al. A systematic review of antibody mediated immunity to coronaviruses: Antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. MedRxiv 2020. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.; Chromikova, V.; McMahon, M.; Jiang, K.; Asthagiri-Arunkumar, G.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. MedRxiv 2020. [Google Scholar] [CrossRef]
- Hsueh, P.R.; Huang, L.M.; Chen, P.J.; Kao, C.L.; Yang, P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004, 10, 1062–1066. [Google Scholar] [CrossRef]
- Haveri, A.; Smura, T.; Kuivanen, S.; Österlund, P.; Hepojoki, J.; Ikonen, N.; Pitkäpaasi, M.; Blomqvist, S.; Rönkkö, E.; Kantele, A.; et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance 2020, 25, 2000266. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Wang, A.; Liu, M.; Wang, Q.; Chen, J.; Xia, S.; Ling, Y.; Zhang, Y.; Xun, J.; Lu, L.; et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. MedRxiv 2020. [Google Scholar] [CrossRef]
- Lou, B.; Li, T.; Zheng, S.; Su, Y.; Li, Z.; Liu, W.; Yu, F.; Ge, S.; Zou, Q.; Yuan, Q.; et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. MedRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. MedRxiv 2020. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020. [Google Scholar] [CrossRef]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The RBD Of The Spike Protein Of SARS-Group Coronaviruses Is A Highly Specific Target Of SARS-CoV-2 Antibodies But Not Other Pathogenic Human and Animal Coronavirus Antibodies. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Reina, J. Remdesivir, the antiviral hope against SARS-CoV-2. Rev. Esp. Quimioter. 2020, 33, 176–179. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef]
- Kruse, R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Research 2020, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Chan, J.F.W.; Azhar, E.I.; Hui, D.S.C.; Yuen, K.-Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Momattin, H.; Dib, J.; Memish, Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: An observational study. Int. J. Infect. Dis. 2014, 20, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H.; Leist, S.R.; Yount, B.L.; McAnarney, E.T.; Graham, R.L.; Waters, K.M.; Baric, R.S. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018, 92, e00710-18. [Google Scholar] [CrossRef]
- Graham, R.L.; Deming, D.J.; Deming, M.E.; Yount, B.L.; Baric, R.S. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun. Biol. 2018, 1, 179. [Google Scholar] [CrossRef]
- Spruth, M.; Kistner, O.; Savidis-Dacho, H.; Hitter, E.; Crowe, B.; Gerencer, M.; Brühl, P.; Grillberger, L.; Reiter, M.; Tauer, C.; et al. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine 2006, 24, 652–661. [Google Scholar] [CrossRef]
- Wang, S.-F.; Tseng, S.-P.; Yen, C.-H.; Yang, J.-Y.; Tsao, C.-H.; Shen, C.-W.; Chen, K.-H.; Liu, F.-T.; Liu, W.-T.; Chen, Y.-M.A.; et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014, 451, 208–214. [Google Scholar] [CrossRef]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Raj, V.S.; Haagmans, B.L. Middle East respiratory syndrome coronavirus vaccines: Current status and novel approaches. Curr. Opin. Virol. 2017, 23, 49–58. [Google Scholar] [CrossRef]
- Sui, J.; Li, W.; Murakami, A.; Tamin, A.; Matthews, L.J.; Wong, S.K.; Moore, M.J.; Tallarico, A.S.C.; Olurinde, M.; Choe, H.; et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 2004, 101, 2536–2541. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Zakhartchouk, A.N.; Sharon, C.; Satkunarajah, M.; Auperin, T.; Viswanathan, S.; Mutwiri, G.; Petric, M.; See, R.H.; Brunham, R.C.; Finlay, B.B.; et al. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: Implications for a subunit vaccine. Vaccine 2007, 25, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Andreadakis, Z.; Kumar, A.; Roman, R.G.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar]
- World Health Organization. Draft Landscape of COVID-19 Candidate Vaccines. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 31 July 2020).
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020. [Google Scholar] [CrossRef]
- Kim, T.W.; Lee, J.H.; Hung, C.-F.; Peng, S.; Roden, R.; Wang, M.-C.; Viscidi, R.; Tsai, Y.-C.; He, L.; Chen, P.-J.; et al. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 4638–4645. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Preliminary Report. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.M.; Vinetz, J.M. Dexamethasone in the management of covid-19. BMJ 2020, 370, m2648. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Jan, J.-T.; Ma, S.-H.; Kuo, C.-J.; Juan, H.-F.; Cheng, Y.-S.E.; Hsu, H.-H.; Huang, H.-C.; Wu, D.; Brik, A.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Chan, K.S.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- World Health Organization. “Solidarity” Clinical Trial for COVID-19 Treatments. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed on 18 March 2020).
- Rajarshi, K.; Chatterjee, A.; Ray, S. Combating COVID-19 with mesenchymal stem cell therapy. Biotechnol. Rep. 2020, 26, e00467. [Google Scholar] [CrossRef]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef]
- Hetta, H.F.; Mekky, M.A.; Khalil, N.K.; Mohamed, W.A.; El-Feky, M.A.; Ahmed, S.H.; Daef, E.A.; Medhat, A.; Nassar, M.I.; Sherman, K.E.; et al. Extra-hepatic infection of hepatitis C virus in the colon tissue and its relationship with hepatitis C virus pathogenesis. J. Med. Microbiol. 2016, 65, 703–712. [Google Scholar] [CrossRef]
- Hetta, H.F.; Mekky, M.A.; Khalil, N.K.; Mohamed, W.A.; El-Feky, M.A.; Ahmed, S.H.; Daef, E.A.; Nassar, M.I.; Medhat, A.; Sherman, K.E.; et al. Association of colonic regulatory T cells with hepatitis C virus pathogenesis and liver pathology. J. Gastroenterol. Hepatol. 2015, 30, 1543–1551. [Google Scholar] [CrossRef]
- Mehta, M.; Hetta, H.F.; Abdel-Hameed, E.A.; Rouster, S.D.; Hossain, M.; Mekky, M.A.; Khalil, N.K.; Mohamed, W.A.; El-Feky, M.A.; Ahmed, S.H.; et al. Association between IL28b rs12979860 single nucleotide polymorphism and the frequency of colonic T reg in chronically HCV-infected patients. Arch. Virol. 2016, 161, 3161–3169. [Google Scholar] [CrossRef]
- Choy, E.H.; De Benedetti, F.; Takeuchi, T.; Hashizume, M.; John, M.R.; Kishimoto, T. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020, 16, 335–345. [Google Scholar] [CrossRef]
- Roll, P.; Muhammad, K.; Schumann, M.; Kleinert, S.; Einsele, H.; Dorner, T.; Tony, H.P. In vivo effects of the anti-interleukin-6 receptor inhibitor tocilizumab on the B cell compartment. Arthritis Rheum. 2011, 63, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Kollias, A.; Kyriakoulis, K.G.; Dimakakos, E.; Poulakou, G.; Stergiou, G.S.; Syrigos, K. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: Emerging evidence and call for action. Br. J. Haematol. 2020, 189, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Zahran, A.M.; Ashmawy, A.M.; Rayan, A.; Elkady, A.; Elsherbiny, N.M.; Hetta, H.F. Frequency and implications of natural killer and natural killer T cells in hepatocellular carcinoma. Egypt. J. Immunol. 2018, 25, 45–52. [Google Scholar]
- Hammer, Q.; Ruckert, T.; Romagnani, C. Natural killer cell specificity for viral infections. Nat. Immunol. 2018, 19, 800–808. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.-M.; Yu, X.-H.; Tang, S.-L.; Tang, C.-K. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int. J. Antimicrob. Agents 2020, 55, 105951. [Google Scholar] [CrossRef]
- Liu, D.; Chen, M.; Mendoza, B.; Cheng, H.; Hu, R.; Li, L.; Trinh, C.T.; Tuskan, G.A.; Yang, X. CRISPR/Cas9-mediated targeted mutagenesis for functional genomics research of crassulacean acid metabolism plants. J. Exp. Bot. 2019, 70, 6621–6629. [Google Scholar] [CrossRef]
- Dorner, T.; Radbruch, A. Antibodies and B cell memory in viral immunity. Immunity 2007, 27, 384–392. [Google Scholar] [CrossRef]
- Liebig, T.M.; Fiedler, A.; Zoghi, S.; Shimabukuro-Vornhagen, A.; von Bergwelt-Baildon, M.S. Generation of human CD40-activated B cells. J. Vis. Exp. 2009, 32, 1373. [Google Scholar] [CrossRef]
- Johnson, M.J.; Laoharawee, K.; Lahr, W.S.; Webber, B.R.; Moriarity, B.S. Engineering of Primary Human B cells with CRISPR/Cas9 Targeted Nuclease. Sci. Rep. 2018, 8, 12144. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Thomsen, A.R. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Case, L.K.; Wang, C.; Yurkovetskiy, L.; Dikiy, S.; Golovkina, T.V. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity 2011, 35, 135–145. [Google Scholar] [CrossRef]
- Carpenito, C.; Milone, M.C.; Hassan, R.; Simonet, J.C.; Lakhal, M.; Suhoski, M.M.; Varela-Rohena, A.; Haines, K.M.; Heitjan, D.F.; Albelda, S.M.; et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. USA 2009, 106, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhao, Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell 2017, 8, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 2019, 76, 826–837. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmood, Z.; Alrefai, H.; Hetta, H.F.; A. Kader, H.; Munawar, N.; Abdul Rahman, S.; Elshaer, S.; Batiha, G.E.-S.; Muhammad, K. Investigating Virological, Immunological, and Pathological Avenues to Identify Potential Targets for Developing COVID-19 Treatment and Prevention Strategies. Vaccines 2020, 8, 443. https://doi.org/10.3390/vaccines8030443
Mahmood Z, Alrefai H, Hetta HF, A. Kader H, Munawar N, Abdul Rahman S, Elshaer S, Batiha GE-S, Muhammad K. Investigating Virological, Immunological, and Pathological Avenues to Identify Potential Targets for Developing COVID-19 Treatment and Prevention Strategies. Vaccines. 2020; 8(3):443. https://doi.org/10.3390/vaccines8030443
Chicago/Turabian StyleMahmood, Zafar, Hani Alrefai, Helal F. Hetta, Hidaya A. Kader, Nayla Munawar, Sheikh Abdul Rahman, Shereen Elshaer, Gaber EI-Saber Batiha, and Khalid Muhammad. 2020. "Investigating Virological, Immunological, and Pathological Avenues to Identify Potential Targets for Developing COVID-19 Treatment and Prevention Strategies" Vaccines 8, no. 3: 443. https://doi.org/10.3390/vaccines8030443
APA StyleMahmood, Z., Alrefai, H., Hetta, H. F., A. Kader, H., Munawar, N., Abdul Rahman, S., Elshaer, S., Batiha, G. E.-S., & Muhammad, K. (2020). Investigating Virological, Immunological, and Pathological Avenues to Identify Potential Targets for Developing COVID-19 Treatment and Prevention Strategies. Vaccines, 8(3), 443. https://doi.org/10.3390/vaccines8030443








