1. Introduction
There are an estimated one billion cases of seasonal influenza reported globally every year. These annual epidemics are estimated to result in about three to five million cases of severe respiratory symptoms, leading to 290,000 to 600,000 deaths [
1,
2]. Vaccination remains the most effective way to control seasonal influenza [
3]. Therefore, the World Health Organization (WHO) recommends annual vaccination against seasonal influenza for healthcare workers and high-risk groups: pregnant women, children aged 6–59 months, the elderly, and persons with specific chronic medical conditions [
4]. Many countries run their own national seasonal influenza immunization programs strictly under WHO guidelines [
5]. However, scientific periodicals still offer insufficient analytical information on the composition of already licensed products on the market. New sophisticated protocols introduced into practice just recently are based on label-free quantitative mass spectrometry. Yet, they have not been used to study commercially available inactivated vaccines. Currently, the hemagglutinin (HA)-antibody level in a human body is evaluated after immunization to determine the efficacy of vaccines. As far as quality control is concerned, it uses several tests, some of which are semi-quantitative in nature. In this regard, impurities such as substrate cell proteins, which are the major causes of side effects at immunization, remain undetectable or are masked by addition of polymers or adjuvants. Their abundance remains uninvestigated. In the Russian Federation, vaccination against influenza is included in the National Immunization Program (NIP) Schedule; NIP vaccines are only supplied by national manufacturers. A relatively small number of vaccines are procured from global manufacturers abroad, i.e., outside the NIP framework. According to the Global Action Plan for Influenza Vaccines (GAP) [
6], the global seasonal influenza vaccine production was 1.467 billion doses in 2015, of which 88% were inactivated influenza vaccines (IIVs) produced through influenza virus replication in the allantoic cavity of chicken embryos (i.e., egg-based vaccines) [
7].
Seasonal influenza vaccines include influenza A viruses, subtypes H1N1 and H3N2, and influenza B viruses, Yamagata and Victoria lineages. Trivalent vaccines include one of the influenza B lineages, and tetravalent vaccines include both influenza B lineages [
8,
9]. However, due to antigenic drift in surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) of the influenza virus, the strains of these types of viruses circulating in the human populations are constantly changing. The WHO annually updates the list of strains recommended for inclusion in seasonal influenza vaccines based on the monitoring of strains circulating in the population in order to maintain the relevance of the antigenic composition of seasonal vaccines [
10].
In accordance with recommendations, licensed laboratories produce candidate vaccine viruses (CVVs), high-growth reassortant viruses adapted for replication in a production substrate and carrying the surface antigens (HA and NA) of the recommended strains. The success of CVV adaptation to replication in chicken embryos may vary significantly depending on the virus strain and production year. Therefore, vaccine manufacturers may face the challenge of insufficient efficiency of vaccine antigen production [
11]. In addition to the growth rate in chicken embryos, CVVs may vary in terms of sensitivity to inactivating and cleaving agents, as well as conditions of virion concentrations and treatment of the virus-containing fluid. All the general steps in the downstream processing of split-virion IIVs (shown in
Table S1) determine the content of HA, other viral proteins, and cellular substrate proteins (i.e., chicken (
Gallus gallus) proteins) within the vaccine, as well as the microstructure of molecular aggregates that are part of the vaccine [
12]. These characteristics, therefore, determine vaccine immunogenicity and safety. Possible variability of the protein composition of commercial IIVs highlights the need for control upon entry into the market (i.e., post-marketing control).
The main idea of the protective immunity elicited by the vaccine comes from production of HA-specific antibodies that inhibit the receptor-binding activity of HA (hemagglutination-inhibiting antibodies) [
13]. Thus, the current WHO guidelines for controlling the final bulk-protein composition (i.e., the step immediately preceding the filling process) of IIVs include three tests: determining the HA, total protein, and ovalbumin contents of each strain of the influenza virus [
14]. Depending on the manufacturing year and the manufacturer, relative amounts of NA, matrix protein (M1), and nuclear protein (NP) (as well as the immune responses induced by them) in different commercially available inactivated split vaccines may vary significantly, from several up to dozens of times [
15]. Thus, new sophisticated platforms for quick and efficient characterization of the manufactured vaccine products should be launched.
Recently, various mass spectrometric approaches for qualitative and quantitative analyses of the proteomes of influenza vaccines have been shown to be feasible. Creskey et al. [
16] showed that liquid chromatography and tandem mass spectrometry (LC-MS/MS) may be used to analyze the amino acid sequences of HA and NA in trivalent inactivated influenza vaccines. Hawksworth et al. [
17] revealed that a quantitative proteomic approach based on label-free quantitation of the relative protein content using the LC-MS/MS assay provided a detailed description of the protein composition of live influenza vaccines. Various mass spectrometric approaches for measuring the absolute concentrations of HA and NA in vaccines have also been developed [
18,
19,
20,
21,
22]. An approach was proposed for measuring the absolute HA concentration in vaccines based on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and a sample-deglycosylation procedure [
23] as an alternative to single radial immunodiffusion (SRID) used for measuring HA concentrations in vaccines during the production process [
14]. Applying the dynamic light scattering (DLS) technique in combination with transmission electron microscopy (TEM) may also provide valuable data for characterization of influenza vaccine microstructures, that is, measuring the hydrodynamic radii of particles and determining the degree of dispersion of vaccine suspensions [
12].
This study is the first that is conducting comparative morphological and proteomic analyses of IIVs licensed in the Russian Federation, the most available on the market and included in the NIP. We tested the applicability and informational content of the label-free LC-MS/MS quantitation of protein content for IIV proteome analysis. As supplementary assays, we used SDS-PAGE of vaccine products, deglycosylated with peptide n-glycosidase F (PNGase F), as well as DLS in combination with TEM. The holistic approach implemented in our study enables us to obtain a necessarily detailed description of the compositions of the studied IIVs quite quickly and independently from reference reagents which could be useful for additional post-marketing control of commercially available inactivated vaccines for prevention of seasonal influenza.
2. Materials and Methods
2.1. IIV Samples
This study covered the inactivated split-virion vaccines (IIVs) for prevention of seasonal influenza licensed in Russia. All studied vaccines were produced by growing influenza viruses in chicken embryos. Both trivalent inactivated influenza vaccines (IIV3) and quadrivalent inactivated influenza vaccines (IIV4) were investigated. The list of vaccines studied is given below:
U3—Ultrix® (IIV3, FORT LLC, Russia);
U4—Ultrix® Quadri (IIV4, FORT LLC, Russia);
SGU—SOVIGRIPP® (IIV3, NPO Microgen JSC, Russia);
SGF—SOVIGRIPP® (IIV3, FORT LLC, Russia);
GP—Grippol® plus, (IIV3, NPO Petrovax Pharm LLC, Russia); and
VG—VAXIGRIP® (IIV3, Sanofi Pasteur С.А., France).
The strain composition of all vaccines was consistent with the WHO recommendations for 2019–2020 seasonal influenza vaccines in the Northern Hemisphere. For trivalent vaccines:
Influenza А virus (H1N1): A/Brisbane/02/2018 (H1N1)pdm09-like virus;
Influenza А virus (H3N2): A/Kansas/14/2017 (H3N2)-like virus; and
Influenza B virus: B/Colorado/06/2017-like virus (B/Victoria/2/87 lineage).
For quadrivalent vaccines—additional strain:
Normative HA content of each subtype of the influenza virus per dose (0.5 mL):
U3: A (H1N1)—15 μg; А (Н3N2)—15 μg; В (Victoria)—15 μg;
U4: A (H1N1)—15 μg; А (Н3N2)—15 μg; В (Victoria)—15 μg; В (Yamagata)—15 μg;
SGU: A (H1N1)—5 μg; А (Н3N2)—5 μg; В (Victoria)—11 μg;
SGF: A (H1N1)—5 μg; А (Н3N2)—5 μg; В (Victoria)—11 μg;
GP: A (H1N1)—5 μg; А (Н3N2)—5 μg; В (Victoria)—5 μg;
VG: A (H1N1)—15 μg; А (Н3N2)—15 μg; В (Victoria)—15 μg.
Vaccines U3, U4, and VG are free of adjuvants and preservatives. Vaccines SGU and SGF contain the SOVIDONТМ synthetic polymer (a copolymer of 2-methyl-5-vinylpyridine and N-vinylpyrrolidone)—500 μg per dose (0.5 mL). Vaccine GP contains the Polyoxidonium® synthetic polymer (high-polymeric units of 100 kDa based on both N-oxide 1.4-ethylene piperazine and (N-carboxyethyl-) 1.4 ethylene piperazine bromide)—500 μg per dose (0.5 mL). As claimed by the vaccine manufacturers, these polymers act as adjuvants. The SGU vaccine also contains thimerosal (merthiolate) as a preservative—50 μg per dose (0.5 mL).
2.2. Lowry Protein Assay
The total protein content in the vaccines was measured using the Lowry protein assay (Peterson’s modification) with protein precipitation [
24] using the Total Protein Kit, Micro Lowry reagent kit (Sigma-Aldrich, Germany, product number: TP0300) in accordance with the manufacturer’s protocol. Optical density was measured at 650 nm on Infinite
® 200 PRO plate reader (TECAN, Switzerland) in 96-well plates (Greiner, Kremsmünster, Austria). Optical density was measured in a 200 μL volume of the test solution per well. The optical density of the solution in each well was determined as the average of 25 measurements. Optical densities of all solutions were measured in three independent repeats, and their arithmetic mean was used to draw the calibration curve.
2.3. Peptide N-Glycosidase F (PNGase F) Treatment
Vaccine samples as they are (without additional concentration) were processed with PNGase F before they were loaded into gels for SDS-PAGE analysis using a deglycosylation kit manufactured by New England Biolabs (NEB), Ipswich, MA, USA (catalog number P0704L) in accordance with the manufacturer’s protocol. In brief, 1× Glycoprotein Denaturing Buffer (from 10× Stock) (NEB) was added to the vaccine samples, heated to 100 °C, incubated at this temperature for 10 min, cooled in ice for about 10 s, and centrifuged at 15,000×
g for 10 s. Then, 1× GlycoBuffer 2 (10×) (NEB), 1× NP-40 (10%) (NEB), and PNGase F (NEB) were added to the reaction mix, the latter at a ratio of 5 activity units per μg of total protein in the sample. The reaction mix was subsequently incubated at 37 °C overnight. The processed samples were loaded onto the gels in equal volumes (see SDS-PAGE procedure in
Section 2.4).
To increase the coverage of surface N-glycosylated influenza virus glycoproteins HA and NA by MS-identified tryptic peptides, we also treated samples with PNGase F before LC-MS/MS analysis according to [
16] with some modifications. For this, aliquots of the vaccine samples containing 10 μg of total protein were selected (based on preliminary quantitation of total protein by the Lowry protein assay, as described in
Section 2.2). The volumes were brought to 100 μL with water (MS grade). Because of the low protein content in the SGU and SGF samples, 10-fold concentrates obtained via centrifugation through 5 kDa molecular weight cut-off filters Vivaspin Turbo 4 (Sartorius, Göttingen, Germany) were used. ProteaseMax (Promega, Madison, WI, USA) 0.05%, acetonitrile 5% (Sigma Aldrich, St. Louis, MO, USA), and triethylammonium bicarbonate 50 mM (Sigma Aldrich, USA) were added to each sample (final concentrations). The reaction mix volume was brought to 200 μL with water, heated to 100 °C and maintained at this temperature for 10 min, then cooled in ice for 10 s, after which 0.1 μL (50 activity units) of PNGase F (NEB) was added to each sample. The reaction mix was then incubated at 37 °C overnight. The reaction mix was then transferred to the sample-preparation stage for LC-MS/MS analysis.
2.4. SDS-PAGE
Denaturing one-dimensional electrophoresis was performed using sodium dodecyl sulfate (SDS)—polyacrylamide gel electrophoresis with a 12.5% concentration of acrylamide (Sigma Aldrich, USA) in the separating gel and 5% concentration in the stacking gel [
25,
26]. Combs with a 5 mm tooth width were used to form pockets for the samples. In order to reduce disulfide bonds, the analyzed samples were placed in the sample buffer (2% lithium dodecyl sulfate (LDS), 0.065 M Tris-HCl (pH 6.8), 1% dithiothreitol (DTT), 10% glycerol, and 0.01% bromophenol blue (all from Sigma Aldrich, USA)) and were heated in a boiling water bath for 2 min. An amount of 1 μL of the Precision Plus Protein Standard (BioRad, Hercules, CA, USA) was loaded as a marker. A maximum of 40 μL (32 μL of the analyzed sample and 8 μL of the buffer) was applied to the lane. Electrophoretic separation (SDS-PAGE) of the sample proteins was performed using the Hoefer miniVE system (Hoefer Inc., Holliston, MA, USA) (gel size of 80 × 90 × 1 mm). Gels were stained with Coomassie Blue R-350 (GE Healthcare, USA) for 2 h.
2.5. LC-MS Quality Control Mix
In order to verify label-free LC-MS/MS-based quantitative analysis to correctly determine the protein ratio in a complex multicomponent mix, we used the LC-MS quality control (QC) mix that was added to each of the studied vaccine samples before the start of mass spectrometric analysis (MSA). It was already shown that QC mix is effective in verifying the label-free LC-MS/MS assessment of protein ratios at different stages of production of live attenuated influenza vaccines [
17]. The LC-MS QC mix that we used included four human proteins (control proteins) and four synthetic stable-isotope-labeled (SIL) peptides, each corresponding to one tryptic peptide from one control protein, isolated from blood plasma: fibrinogen (equimolar mix of three subunits FIBA, FIBB, FIBG (in our study, we focused on two, FIBB and FIBG) bound in the native protein by disulfide bonds that break down under reducing conditions), apolipoprotein A1 (APOA), and apolipoprotein B (APOB) (
Table S2). Lyophilized preparations of these proteins were purchased from IMTEK (Russia). Next, we a used high-resolution (HR) liquid chromatography-mass spectrometry (LC-MS) assay to measure the absolute concentration of each control protein in accordance with the approach described by Guo et al. [
22] (see
Section 2.8). The obtained ratios of absolute concentrations of the control proteins were compared with ratios of their abundance obtained via label-free LC-MS/MS-based calculations using the intensity-based absolute-quantitation (iBAQ) algorithm [
27].
The lyophilized proteins were dissolved in MS-grade water to a 2 mg/mL concentration. The resulting stock solutions were stored at –20 °C. Then, protein solutions were quantitatively added to each sample of the studied vaccines after PNGase F treatment and before the reduction/alkylation step (the volume of each sample contained 10 μg of the total vaccine protein and did not exceed 100 μL). Concentrations of control proteins in the vaccine samples were selected such that the minimal concentration of the control protein (APOB) was about 10 times lower than the total HA normative concentration, and the maximum (APOA) was several times higher than the total HA normative concentration (
Table 1). Thus, the concentration range covered by the control proteins was ~1.5 to 2 orders of magnitude. It included the concentrations of the target vaccine proteins.
Selection, synthesis, and determination of the concentrations of SIL peptides were carried out by the Human Proteome Center, Institute of Biomedical Chemistry (IBMC), Moscow, Russia, in accordance with the method described by Kopylov et al. [
28]. Concentrations of synthetic peptides were measured using fluorescent-signal detection of the amino acid derived after peptide acidic hydrolysis. SIL peptides were added in known concentrations to the test vaccine samples one hour before trypsin digestion (see trypsinolysis conditions in
Section 2.6). Concentrations of SIL peptides in the vaccine samples were selected such that they did not differ from standard protein concentrations by more than two orders of magnitude (
Table 1).
2.6. Reduction, Alkylation, and Trypsin Digestion
The sample was prepared for MSA in accordance with the filter-aided sample preparation (FASP) protocol [
29]. In order to reduce and alkylate disulfide bonds, samples were incubated in 4 mM tris (2-carboxyethyl) phosphine (TCEP; Sigma Aldrich, USA) and 6.2 mM 2-chloroacetamide (CAA; Sigma Aldrich, USA) at 80 °С for 30 min. Then, the reaction mix was placed in Microcon YM-10 centrifugal filter units (Merck Millipore, Burlington, MA, USA) and centrifuged at 11,000×
g for 15 min in a thermostatic centrifuge at 20 °C. The samples were rinsed three times by adding 200 μL of the buffer containing 50 mM of tetraethylammonium bicarbonate (pH = 8.5), followed by centrifugation at 11,000×
g for 15 min in a thermostatic centrifuge at 20 °C.
To hydrolyze proteins of the vaccine preparations after the last rinsing, 50 μL of the buffer containing 50 mM tetraethylammonium bicarbonate (pH = 8.5) and trypsin (Promega, USA) with an enzyme/total protein weight ratio of 1/50 were added to the samples. The mix was incubated overnight at 37 °C, with 350 rpm shaking. Samples were centrifuged on filters at 11,000× g for 15 min in a thermostatic centrifuge at 20 °C to obtain a peptide solution. Then, the filters were rinsed with 50 μL of a 30% solution of formic acid (Sigma Aldrich, USA) by centrifugation at 11,000× g for 15 min in a thermostatic centrifuge at 20 °C. The filtrate was dried in a vacuum concentrator and dissolved in 20 μL of 5% formic acid for subsequent MSA.
2.7. LC-MS/MS
LC-MS/MS was performed using the Ultimate 3000 RSLCnano chromatographic HPLC system (Thermo Scientific, Waltham, MA, USA) coupled with a Q Exactive HF-X mass spectrometer (Thermo Scientific, USA). We loaded 1 μg of the peptide mix onto the Acclaim μ-Precolumn enrichment column (0.5 × 3 mm, 5 μm particle size; Thermo Scientific, USA) at a flow rate of 10 μL per minute for four minutes in isographic mode using buffer C as a mobile phase (2% acetonitrile (Sigma Aldrich, USA), 0.1% formic acid in deionized water). Peptides were then separated on the Acclaim Pepmap® C18 HPLC column (75 μm × 150 mm, 2 μm particle size; Thermo Scientific, USA) in gradient-elution mode. The gradient was formed by mobile phases A (0.1% formic acid) and B (80% acetonitrile, 0.1% aqueous solution of formic acid) at a flow rate of 0.3 μL per minute. The column was rinsed with 2% mobile phase B for four minutes, with subsequent a linear increase in mobile phase B concentration to 35% in 45 min and a linear increase in phase B concentration to 99% in 5 min; after 10 min rinsing with 99% buffer B, the concentration of this buffer was linearly reduced to the initial level of 2% in six minutes. Overall, the analysis took 70 min.
MSA was performed on a Q Exactive HF-X mass spectrometer (Thermo Scientific, USA) in positive-ionization mode using the NESI (nanoelectrospray) source. The MSA was performed under 2.1 kV emitter voltage and 240 °C capillary temperature. Panoramic scanning was carried out in a mass range of 300 to 1500 m/z with a 120,000 resolution. In tandem scanning, resolution was set to 15,000 in a 100 m/z mass range to the upper limit, which was automatically determined based on the precursor mass, but of no more than 2000 m/z. Precursor-ion isolation was performed in a window of ±1 Da. The maximum number of ions allowed for isolation in the MS2 mode was set to be within 40. In addition, 50,000 units was set as the cut-off limit for selecting the precursor for tandem analysis, while the normalized collision energy (NCE) was equal to 29. Only ions with charges from z = 2+ to 6+ were considered for tandem scanning. Maximum accumulation time was 50 ms for precursor ions and 110 ms for fragment ions. The AGC (automatic gain control) value for precursors and fragment ions was set to 1 × 106 and 2 × 105, respectively. All measured precursors were dynamically excluded from tandem MS/MS analysis for 90 s. Each sample was analyzed in three runs, and the results were used for further bioinformatics processing.
2.8. Data Processing
The densitometry of protein bands was determined using ImageMaster™ 2D Platinum 7.0 software (GE Healthcare, Chicago, IL, USA). Molecular-weight values for protein bands were determined using Precision Plus Protein Standards (BioRad, Hercules, CA, USA). The relative volume of the protein spot (% vol) value was used to assess protein weight in the band on the gel. To evaluate the relationship between total HA weight and total protein weight in the vaccine (relative HA weight, %), we summed the % vol of the HA1 and HA2 bands (after sample N-deglycosylation) on the lane and divided the value by the sum of the % vol of all bands of the same lane. The absolute weight of total HA was calculated as the relative HA weight of the total protein weight measured by the Lowry protein assay (with protein precipitation).
Mass spectra were analyzed using MaxQuant 1.5.8.3 software [
30] in accordance with the Hutchinson and Stegmann protocol [
31]. Files with the mass-spectrometric analysis results for all three runs for each sample were analyzed together using standard settings and the following parameters. Enzyme, trypsin/P; variable modifications, oxidation (M) and acetyl (Protein N-ter); fixed modifications, carbamidomethyl (C).
Peptide spectra were matched to a database containing the reference chicken (
Gallus gallus) proteome (UniProt database UP000000539 (
https://www.uniprot.org/proteomes/UP000000539)) and the expected proteomes of virions of the influenza virus strains recommended by the WHO for inclusion in 2019–2020 seasonal influenza vaccines in the Northern Hemisphere. The lists of expected virion proteins were compiled based on recent studies on mass spectrometric analysis of influenza virions [
32] and live attenuated influenza vaccines [
17]. Viral protein sequences for inclusion in the database were consensus sequences generated by a computer algorithm (Geneious Prime 2019.8.1.9 (Biomatters, Auckland, New Zealand)) on the basis of multiple alignments of amino acid sequences of viral proteins of the isolates of influenza virus strains with completely sequenced genomes uploaded to the EpiFlu
® GISAID public database [
17,
33]. We also included PNGase F, trypsin, and human protein sequences in our database from the LC-MS QC mix. All proteins belonging to the automatically generated base of decoy proteins (reverse sequences) were excluded from the identified proteins.
The iBAQ value was used for label-free quantitation of protein abundance [
27]. For the quantitation of vaccine proteomes, all mass spectrometrically identified proteins were grouped in accordance with genes encoding these proteins, namely, HA A, HA B, NA, NP, M1, M2, non-structural protein one (NS1), nuclear export protein (NEP), three viral polymerase subunits - PB1, PB2, PA, and NB. Justification of the viral-protein grouping principle is given in
Section 3.3.
Absolute concentrations of total HA in the vaccines were calculated using label-free LC-MS/MS analysis and based on the fact that the iBAQ value of the protein was directly proportional to the amount of substance of this protein. The proportionality factor was the same for all proteins from one sample [
27]. The algorithm for calculating the HA concentration was similar to that based on the densitometry results. First, we calculated the relative weight of total HA (relative HA weight, %) as the ratio of the sum of products of the HA iBAQ of each strain multiplied by their molecular weights to the sum of products of the iBAQ of all proteins in the sample multiplied by their molecular weights:
where Σ is the summation symbol;
, HA iBAQ is the value of one influenza virus strain of the studied sample; I the is influenza virus-strain number;
is the molecular weight of influenza virus strain HA (molecular weights of deglycosylated HA were used);
, iBAQ is the value of the protein of the studied sample; and k is the protein number of the studied sample. Absolute concentration of total HA was calculated using the following formula:
where total protein (μg/mL) was measured via the Lowry protein assay.
Absolute concentrations of control proteins were determined by the corresponding SIL peptides by the (HR) LC-MS assay in accordance with the protocol validated by Guo et al. [
22]. Extracted ion chromatograms of natural tryptic peptides and corresponding SIL peptides were generated. Chromatograms were analyzed using XCalibur 3.1 software (Thermo Scientific, USA) supplied with the mass spectrometer. The following settings were used for automatic peak detection and integration: peak-integration algorithm, ICIS; smooth points, 11; baseline window, 100; minimal peak height (S/N), 3; mass tolerance, 5 ppm. Ratios of areas under the peaks of the corresponding natural and synthetic peptides were interpreted as the ratios of their concentrations. As was shown in the study by Guo et al. [
22], one representative peptide is sufficient for accurate measurement of protein concentration. Thus, concentrations of tryptic peptides obtained were equated with concentrations of the starting proteins from which these tryptic peptides were derived.
2.9. Dynamic Light Scattering (DLS)
DLS particle-size distributions in the samples were measured by means of a Zetasizer Nano ZS (Malvern Panalytical, Malvern, UK) at a fixed scattering angle of 173° in a thermostatic cell at 25 °C. We used 100 averages of 25 s scans to obtain a histogram with ~5% accuracy. Data were analyzed using Dispersion Technology software, version 5.10 (Dispersion Technology, Inc., Bedford Hills, NY, USA).
2.10. Negative-Stain Transmission Electron Microscopy
Vaccine solutions were deposited on formvar–carbon-coated grids (TED Pella, Redding, CA, USA), incubated for two minutes. After that, excess solution was removed, and the samples were then stained for 20 s with a 2% water solution of phosphotungstic acid (PTA; Sigma Aldrich, USA), pH 7.0. Samples were viewed with a transmission electron microscope Jeol JEM-2100 (JEOL Ltd., Tokyo, Japan).
4. Discussion
Vaccination still remains the best protection currently available against seasonal influenza [
3]. Among the many types of seasonal influenza vaccines, the most widely used are inactivated influenza vaccines (IIVs) based on influenza virus propagation in chicken embryos [
7]. Nevertheless, there are differences in the processes used by manufacturers to obtain the final product, especially at the multistep stage of downstream processing [
12]. Annual updates of the WHO recommendations on influenza virus strains to be used in seasonal influenza vaccines and shortened production periods create additional difficulties in maintaining the consistency of vaccine formulations from year to year [
11]. The standards for controlling the composition of inactivated influenza vaccines used today depend on the presence of reference reagents; they consider only a few key components, and do not allow for complete characterization of influenza vaccine composition [
14]. Possible undetectable variations in the complex composition of inactivated influenza vaccines may affect the safety, immunogenicity, and ultimately the efficacy of these vaccines.
To address the above challenges, this paper first describes a comparative analysis of commercially available inactivated seasonal influenza vaccines (IIVs) included in the Russian National Immunization Program schedule. It studies the proteomes and molecular morphology of vaccines using a holistic approach including such methods as SDS-PAGE and sample deglycosylation, LC-MS/MS, and dynamic light scattering in combination with transmission electron microscopy. The methods used do not depend on reference reagents and therefore allowed for a quick and informative post-marketing study of IIVs.
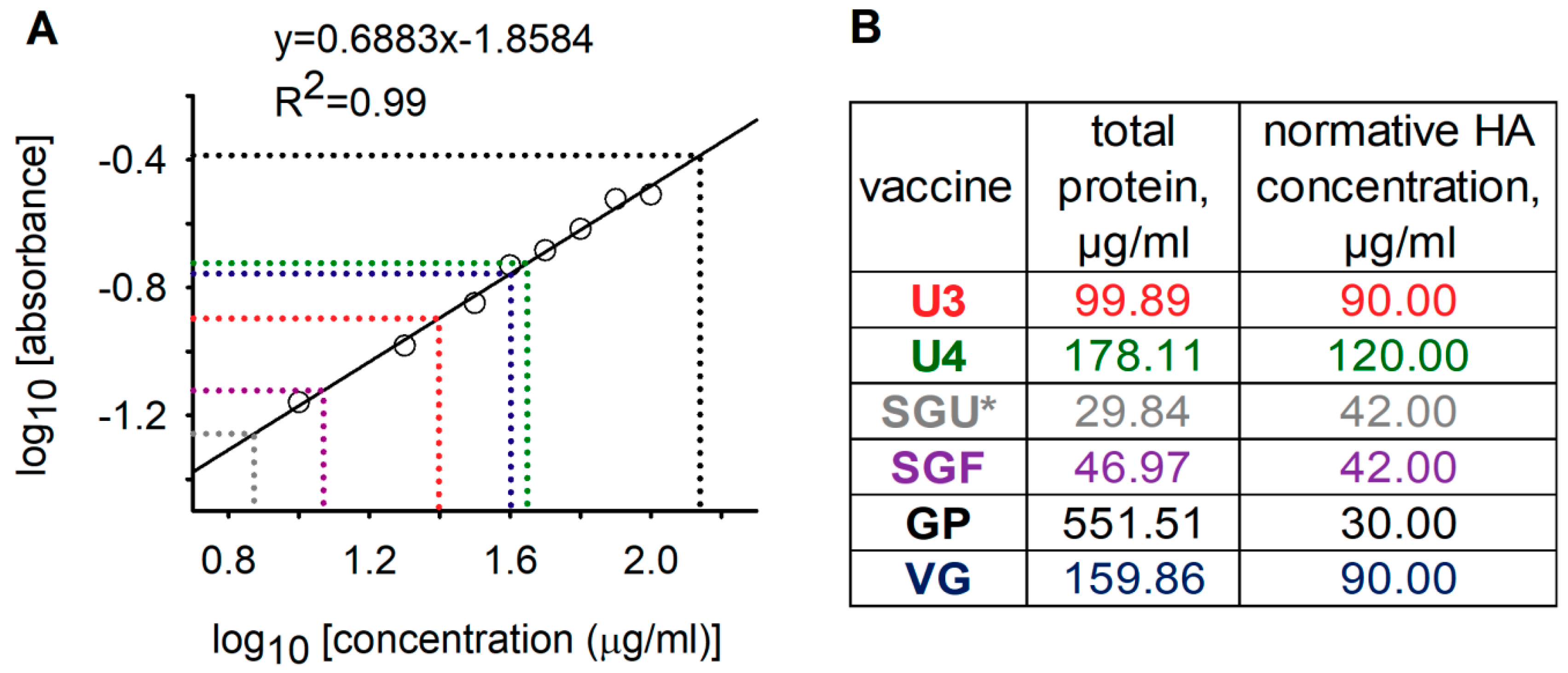
For the general qualitative and quantitative characterization of the protein profiles of IIVs, we used standard methods: total protein quantitation by Lowry protein assay with precipitation and the SDS-PAGE assay. These methods for analysis of vaccines licensed in Russia were shown to have limited applicability. Synthetic polymers present in the SGU and SGF vaccines (SOVIDON
ТМ: a copolymer of 2-methyl-5-vinylpyridine and N-vinylpyrrolidone) and GP (Polyoxidonium
®: high-polymeric units of 100 kDa based on both N-oxide 1.4-ethylene piperazine and (N-carboxyethyl-) 1.4 ethylene piperazine bromide) interfered with total protein quantitation by Lowry protein assay (
Figure 1) and protein separation by SDS-PAGE (
Figure 2).
In vaccines without synthetic polymers (U3, U4, and VG), we revealed similar patterns of electrophoretic migration of proteins in gels. Major protein bands on the gels obtained corresponded in terms of molecular weight with influenza virion proteins: HA (cleaved into HA1 and HA2 subunits after reduction), NP, and M1 (
Figure 2). Thus, we reproduced the results of previous studies based on SDS-PAGE analysis of inactivated influenza vaccines [
12,
35] and purified influenza virus virions [
23,
32] in relation to IIVs selected for study in this paper.
Since the influenza virus HA is an intensely N-glycosylated glycoprotein, the peptide N-glycosidase F (PNGase F) treatment of vaccine samples before application on gels leads to carbohydrate cleavage from this protein and consequently to increased electrophoretic mobility of HA1 and HA2, and to decreased fuzziness of the respective protein bands [
15]. After deglycosylation, HA1 and HA2 bands may be reliably separated from NP and M1, respectively. This fact was used in a number of studies for absolute quantitation of HA content in vaccines using SDS-PAGE assay in combination with densitometry [
11,
15,
26]. In particular, Li et al. [
35] showed that this approach yielded a result that deviated from SRID by 12% to 22%.
In this paper, the PNGase F treatment of vaccine samples expectedly increased electrophoretic mobility of HA1 and HA2 (
Figure 2). Moreover, after deglycosylation of HA1, the subunits formed several bands within the 37 to 45 kDa range. This may have been caused by the presence of several HA1 proteoforms corresponding to different types and subtypes of HA, as well as heterogeneity of HA glycosylation described in previous studies [
43].
Using the results of gel densitometry after deglycosylation (
Figure S1, and
Tables S3 and S4) and our measured total protein concentrations (
Figure 1), we calculated total HA concentrations in the vaccines without synthetic polymers. The approach used was that developed by Harvey et al. [
23]. The obtained total HA concentrations in the vaccines without synthetic polymers ranged from 69.23% to 75.71% of the normative values specified in instructions for the preparations (
Table 2). HA concentrations quantified by SDS-PAGE in our study could have been underestimated due to incomplete deglycosylation and presence of proteins with molecular weights beyond the studied molecular weight range. This indicated the need to optimize the deglycosylation conditions and to select conditions for SDS-PAGE to accurately quantify the HA content by SDS-PAGE. However, this was not among the aims of our study.
For deeper and more sensitive analysis of IIV protein composition, we used the LC-MS/MS assay. As has been shown in a number of studies, this assay provides an effective platform for identification and quantitative analysis of proteins in isolated influenza virus virions and influenza vaccines [
16,
17,
32,
44]. In addition, LC-MS/MS that was based on other physical processes than the Lowry assay and SDS-PAGE, theoretically allowed us to overcome the inapplicability of the Lowry assay and SDS-PAGE for the study of vaccines containing high-molecular-weight artificial polymers (SGU, SGF, GP).
The accuracy of label-free LC-MS/MS-based quantitation of protein concentrations in a complex mix is much lower than that of targeted approaches: mass spectrometry using synthetic isotope-labeled peptides (isotope dilution mass spectrometry (IDMS), high-resolution (HR) LC-MS) [
18,
19,
22], and serological methods SRID [
45] and ELISA (enzyme-linked immunosorbent assay) [
46]. However, in comparison with the targeted methods, label-free LC-MS/MS did not depend on a potentially long and expensive stage of preparation of special reagents for quantitation of specific proteins (i.e., SIL peptide synthesis with a specific amino acid sequence and production of specific antibodies). Therefore, LC-MS/MS is much more appropriate for post-marketing control of IIVs given the annual update of their strain composition and, accordingly, annual update of control reagents. Although this approach is quite expensive today, we hope that it will become cheaper in the near future, as mass spectrometry equipment will become more available in many protein research laboratories.
In a paper recently published by Hawksworth et al. [
17], internal protein standards were used to verify label-free protein quantitation (iBAQ) [
27] of substances representing different stages of downstream processing in live-influenza vaccine production. Following this approach, we used internal standards to verify iBAQ-based protein quantitation in commercially available IIVs. The procedure used in our study was based on measurement of absolute protein concentrations using high-resolution (HR) LC-MS [
22]. Verification of the iBAQ-based evaluation allowed us to perform reliable quantitative proteomic analysis of IIVs. In particular, we could evaluate the relative content of cell substrate proteins in the vaccines (chicken (
Gallus gallus)), and HA A and B, NA, M1, NP, and other structural proteins of influenza viruses. In additionally demonstrating the correctness of the iBAQ-based assessment of the HA content in IIVs, we showed that, in vaccines without synthetic polymers (U3, U4, VG), the total HA content quantified using the LC-MS/MS assay correlated with SDS-PAGE-based measurements (
Table 4).
Host cell proteins are an unavoidable component of any viral vaccine. Complete removal of substrate proteins is impossible not only due to technical limitations of the methods for purification of the virus-containing liquid, but also because of incorporation of host proteins into the structure of the influenza virus virion [
32]. However, downstream processing of IIVs (
Table S1) is aimed at eliminating substrate proteins (chicken proteins) from a final product. Removal of chicken proteins is required to prevent exceeding the maximal level of total protein (no more than 300 μg per dose) [
14] and to minimize the concentration of the potential allergen, ovalbumin (no more than 1 μg per dose) [
46,
47] in the final product. We found that, in the U3, U4, SGU, SGF, and VG vaccines, the relative proportion of chicken proteins (by substance) varied from 18% (SGU) to 36% (SGF). The GP vaccine contained 80% of chicken protein, falling out of said interval, which may raise concerns about the safety of this vaccine compared to the rest of the vaccines studied.
Downstream processing of the virus-containing allantoic fluid (VCAF;
Table S1) in the IIV production process is usually designed to enrich the preparation with the main surface glycoprotein of the influenza virus HA [
12]. This is required to guarantee the vaccine’s ability to stimulate a sufficiently high level of neutralizing antibodies to HA and to form sterilizing immunity as a result. From this point of view, abundance of non-HA (or other) viral proteins, in comparison with the abundance of HA, reflects the success of a strategy for vaccine enrichment with hemagglutinin. However, much of the currently available experimental evidence was on the role of non-HA proteins in protective immunity against influenza [
15]. Considering this, one of the aims of our study was to quantify non-HA viral proteins in different IIVs. This issue is also important in light of the fact that existing WHO recommendations on the composition and quality control of influenza vaccines require manufacturers to control only the HA content (among all viral proteins), setting SRID as a control method [
14]. This method, in addition to its dependence on reference reagents, is difficult to adapt for studies involving low-represented proteins in IIVs, such as NA and other viral proteins. Therefore, the LC-MS/MS assay represents an alternative to SRID, allowing for the use of reference reagents to detect low-presenting proteins in IIVs and, thereby, obtain more detailed information about vaccine composition.
We could reliably detect NA, M1, and NP proteins in all the studied IIVs. Relative abundance of NA varied slightly between the vaccines. A significant scatter was found in the M1 and NP contents of different preparations. This finding is consistent with results of non-HA protein quantitation in commercially available vaccines using Western blot analysis. Differences in the non-HA protein concentration correlate with the intensities of cellular and humoral immune responses [
15]. Thus, our study indicated the potential variability of commercially available IIVs licensed in Russia by their ability to elicit an immune response to non-HA proteins of the influenza virus, and, therefore, potentially stimulate development of cross-protective immunity against various types and subtypes of the influenza virus.
It is worth mentioning that not only proteins but also other biological molecules, e.g., nucleic acids or lipids may affect the vaccine efficiency, boost unspecific immune response, or induce undesirable side-effects [
8,
48]. That is why a comprehensive investigation of modern vaccine preparations using new sophisticated methodologies is of crucial importance. At the moment, the quality assurance/quality control protocols applied for IIV in Russia do not include testing for RNA (e.g., by applying reverse transcription polymerase chain reaction, RT-PCR). Yet, future progress in vaccinology science as well as the demands of vaccinology practice will definitely allow one to provide a comprehensive control of different constituents within the vaccine preparations to understand the responsible factors better.
Dynamic light scattering was used to measure the vaccine-particle diameter and to assess vaccine-suspension uniformity. According to Kon et al. [
12], particle size distribution in split virion IIVs was most affected by the virion cleavage stage (increasing vaccine preparation dispersion) and the subsequent sterile filtration stage (decreasing preparation dispersion and removal of the coarse-particle fraction (≥200 nm)). Therefore, the significant differences in particle distribution found in our vaccines suggest there are differences in the raw virus-containing fluid-processing technologies used by vaccine manufacturers (
Figure 7).
Electron microscopy showed that particles detected by dynamic light scattering were mainly molecular aggregates containing HA (rosettes). As shown by McCraw et al. [
40], HA aggregation into rosettes occurs through hydrophobic interaction among HA transmembrane domains. The presence of HA rosettes in split virion IIVs, similar to those in purified recombinant HA preparations, is understandable in terms of a typical production strategy for this type of vaccine [
12]. During the vaccine production process, the influenza virus is cleaved by detergents (for example, octyl glycoside or Triton X-100), destroying the lipid membrane of the virus and transferring the integrated proteins (HA, NA, M2, NB) into the solution. During downstream processing, the concentration of detergents is critically reduced as a result of fractionation and dilution of the liquid containing inactivated cleaved virions. Removal of detergents from the solution can theoretically lead to protein aggregation through hydrophobic interprotein interactions. Experimental results obtained by Koroleva et al. [
15] also agree with this assumption. Using coimmunoprecipitation, the authors detected aggregation of heterologous HA proteins, as well as M1 and NA proteins with HA protein in licensed split virion IIVs. Thus, the abundance of molecular aggregates we observed in the studied vaccines is consistent with published results (
Figure 6).