Role of the DNA-Binding Protein pA104R in ASFV Genome Packaging and as a Novel Target for Vaccine and Drug Development
Abstract
:1. Introduction
2. ASFV Genome Replication and Packaging
2.1. Viral DNA-Packaging Proteins
2.2. ASFV Genome Structure
2.3. ASFV Structural Proteins and Proteins Involved in Assembly
| ORF | Description | Localization | Reference(s) |
|---|---|---|---|
| B646L | Major capsid protein p72 | capsid | [30,39] |
| B438L | Protein p49 | capsid | [40] |
| E120R | Protein p14.5 | capsid | [33] |
| D117L | Major transmembrane protein p17 | inner envelope | [32,41] |
| E183L | Transmembrane protein pE183L | inner envelope | [42,43] |
| CP2475L | Protein p5, polyprotein pp220 derived | core shell | [44] |
| CP2475L | Protein p14, polyprotein pp220-derived | core shell | [31,44,45,46] |
| CP2475L | Protein p34, polyprotein pp220-derived | core shell | [31,44,45,46] |
| CP2475L | Protein p37, polyprotein pp220-derived | core shell | [31,44,45,46] |
| CP2475L | Protein p150, polyprotein pp220-derived | core shell | [31,44,45,46] |
| CP530R | Protein p8, polyprotein pp62-derived | core shell 1 | [47] |
| CP530R | Protein p15, polyprotein pp62-derived | core shell | [31,45,48] |
| CP530R | Protein p35, polyprotein pp62-derived | core shell | [31,45,48] |
| S273R | Polyprotein processing protease | core shell | [49,50] |
| A104R | Histone-like DNA-binding protein | nucleoid | [34,44] |
| K78R | DNA-binding protein p10 | nucleoid | [51] |
| P1192R | Topoisomerase II | ND | [36,37] |
| B354L | A32L ATPase 2 | ND | [35] |
| D345L | Lambda-like recombinase 2 | ND | [17] |
3. The DNA Binding Protein pA104R
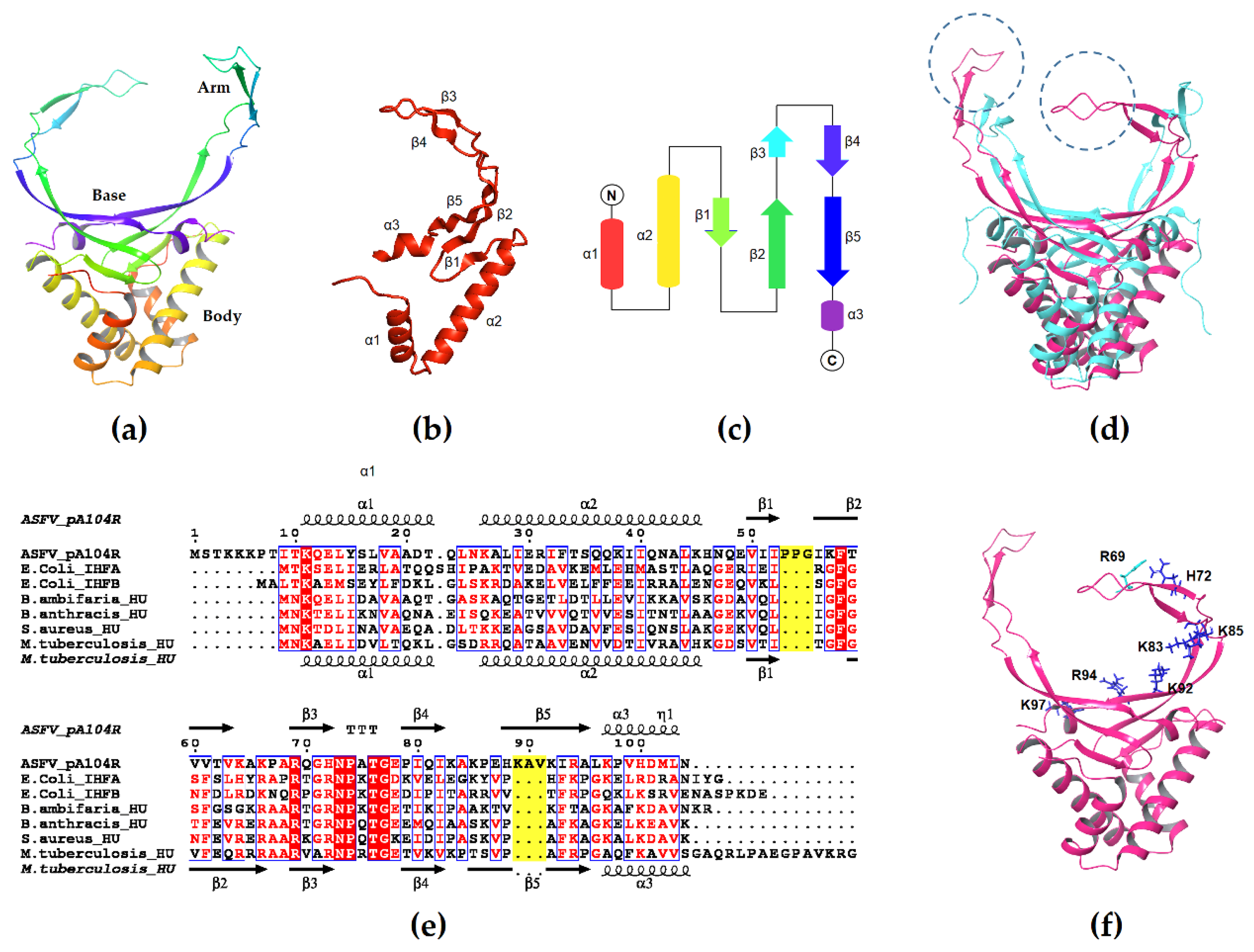
3.1. Structure of pA104R
3.2. Role of pA104R in Viral Packaging
3.3. Phylogenetic Analysis of pA104R
3.4. Potential of pA104R as a Target for Vaccine and Drug Development
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Costard, S.; Mur, L.; Lubroth, J.; Sanchez-Vizcaino, J.M.; Pfeiffer, D.U. Epidemiology of African swine fever virus. Virus Res. 2013, 173, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Global Situation of African Swine Fever. In African Swine Fever (ASF); Report N°47: 2016–2020; WAHIS: Paris, France, 2020. [Google Scholar]
- World Organisation for Animal Health. ASF Situation. In African Swine Fever (ASF); Report N°52: 21 August–3 September 2020; WAHIS: Paris, France, 2020. [Google Scholar]
- Chapman, D.A.G.; Darby, A.C.; Da Silva, M.; Upton, C.; Radford, A.D.; Dixon, L.K. Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus. Emerg. Infect. Dis. 2011, 17, 599–605. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Animal Health and Welfare. Scientific Opinion on African swine fever. EFSA J. 2014, 12, 3628. [Google Scholar] [CrossRef]
- Swinger, K.K.; Rice, P.A. IHF and HU: Flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004, 14, 28–35. [Google Scholar] [CrossRef]
- Sandman, K.; Reeve, J.N. Archaeal chromatin proteins: Different structures but common function? Curr. Opin. Microbiol. 2005, 8, 656–661. [Google Scholar] [CrossRef]
- De Souza, R.F.; Iyer, L.M.; Aravind, L. Diversity and evolution of chromatin proteins encoded by DNA viruses. Biochim. Biophys. Acta 2010, 1799, 302–318. [Google Scholar] [CrossRef] [Green Version]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Chevignon, G.; Thézé, J.; Cambier, S.; Poulain, J.; Da Silva, C.; Bézier, A.; Musset, K.; Moreau, S.J.M.; Drezen, J.-M.; Huguet, E. Functional annotation of Cotesia congregata bracovirus: Identification of viral genes expressed in parasitized host immune tissues. J. Virol. 2014, 88, 8795–8812. [Google Scholar] [CrossRef] [Green Version]
- Michel, B.; Fournier, G.; Lieffrig, F.; Costes, B.; Vanderplasschen, A. Cyprinid herpesvirus 3. Emerg. Infect. Dis. 2010, 16, 1835–1843. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 2010, 53, 284–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, G.; Blanc-Mathieu, R.; Song, C.; Kayama, Y.; Mochizuki, T.; Murata, K.; Ogata, H.; Takemura, M. Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water. J. Virol. 2019, 93, e02130-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelikani, V.; Ranjan, T.; Zade, A.; Shukla, A.; Kondabagil, K. Genome Segregation and Packaging Machinery in Acanthamoeba polyphaga Mimivirus Is Reminiscent of Bacterial Apparatus. J. Virol. 2014, 88, 6069–6075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelikani, V.; Ranjan, T.; Kondabagil, K. Revisiting the genome packaging in viruses with lessons from the “Giants”. Virology 2014, 466–467, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Iyer, L.M.; Balaji, S.; Koonin, E.V.; Aravind, L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006, 117, 156–184. [Google Scholar] [CrossRef] [Green Version]
- Guo, P.; Zhao, Z. Biomotors: Linear, Rotation, and Revolution Motion Mechanisms, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781351136068. [Google Scholar]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Blasco, R.; Agüero, M.; Almendral, J.; Viñuela, E. Variable and constant regions in african swine fever virus DNA. Virology 1989, 168, 330–338. [Google Scholar] [CrossRef]
- González, A.; Talavera, A.; Almendral, J.M.; Viñuela, E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986, 14, 6835–6844. [Google Scholar] [CrossRef] [Green Version]
- Chapman, D.A.G.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89, 397–408. [Google Scholar] [CrossRef]
- Yáñez, R.J.; Rodríguez, J.M.; Nogal, M.L.; Yuste, L.; Enríquez, C.; Rodriguez, J.F.; Viñuela, E. Analysis of the Complete Nucleotide Sequence of African Swine Fever Virus. Virology 1995, 208, 249–278. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.M.; Salas, M.L. African swine fever virus transcription. Virus Res. 2013, 173, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Michaud, V.; Randriamparany, T.; Albina, E. Comprehensive Phylogenetic Reconstructions of African Swine Fever Virus: Proposal for a New Classification and Molecular Dating of the Virus. PLoS ONE 2013, 8, e69662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malogolovkin, A.; Kolbasov, D. Genetic and antigenic diversity of African swine fever virus. Virus Res. 2019, 271, 197673. [Google Scholar] [CrossRef] [PubMed]
- Krug, P.W.; Holinka, L.G.; O’Donnell, V.; Reese, B.; Sanford, B.; Fernandez-Sainz, I.; Gladue, D.P.; Arzt, J.; Rodriguez, L.; Risatti, G.R.; et al. The Progressive Adaptation of a Georgian Isolate of African Swine Fever Virus to Vero Cells Leads to a Gradual Attenuation of Virulence in Swine Corresponding to Major Modifications of the Viral Genome. J. Virol. 2015, 89, 2324–2332. [Google Scholar] [CrossRef] [Green Version]
- De Villiers, E.P.; Gallardo, C.; Arias, M.; da Silva, M.; Upton, C.; Martin, R.; Bishop, R.P. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 2010, 400, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- García-Escudero, R.; Andrés, G.; Almazán, F.; Viñuela, E. Inducible gene expression from African swine fever virus recombinants: Analysis of the major capsid protein p72. J. Virol. 1998, 72, 3185–3195. [Google Scholar] [CrossRef] [Green Version]
- Andrés, G.; Alejo, A.; Salas, J.; Salas, M.L. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 2002, 76, 12473–12482. [Google Scholar] [CrossRef] [Green Version]
- Suárez, C.; Gutiérrez-Berzal, J.; Andrés, G.; Salas, M.L.; Rodríguez, J.M. African swine fever virus protein p17 is essential for the progression of viral membrane precursors toward icosahedral intermediates. J. Virol. 2010, 84, 7484–7499. [Google Scholar] [CrossRef] [Green Version]
- Andrés, G.; García-Escudero, R.; Viñuela, E.; Salas, M.L.; Rodríguez, J.M. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J. Virol. 2001, 75, 6758–6768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frouco, G.; Freitas, F.B.; Coelho, J.; Leitão, A.; Martins, C.; Ferreira, F. DNA-Binding Properties of African Swine Fever Virus pA104R, a Histone-Like Protein Involved in Viral Replication and Transcription. J. Virol. 2017, 91, e02498-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, L.M.; Aravind, L.; Koonin, E. V Common Origin of Four Diverse Families of Large Eukaryotic DNA Viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baylis, S.A.; Dixon, L.K.; Vydelingum, S.; Smith, G.L. African swine fever virus encodes a gene with extensive homology to type II DNA topoisomerases. J. Mol. Biol. 1992, 228, 1003–1010. [Google Scholar] [CrossRef]
- Garcia-Beato, R.; Freije, J.M.P.; López-Otín, C.; Blasco, R.; Viñuela, E.; Salas, M.L. A gene homologous to topoisomerase II in African swine fever virus. Virology 1992, 188, 938–947. [Google Scholar] [CrossRef]
- Coelho, J.; Martins, C.; Ferreira, F.; Leitão, A. African swine fever virus ORF P1192R codes for a functional type II DNA topoisomerase. Virology 2015, 474, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Hingamp, P.M.; Leyland, M.L.; Webb, J.; Twigger, S.; Mayer, R.J.; Dixon, L.K. Characterization of a ubiquitinated protein which is externally located in African swine fever virions. J. Virol. 1995, 69, 1785–1793. [Google Scholar] [CrossRef] [Green Version]
- Epifano, C.; Krijnse-Locker, J.; Salas, M.L.; Salas, J.; Rodríguez, J.M. Generation of filamentous instead of icosahedral particles by repression of African swine fever virus structural protein pB438L. J. Virol. 2006, 80, 11456–11466. [Google Scholar] [CrossRef] [Green Version]
- Simón-Mateo, C.; Freije, J.M.; Andrés, G.; López-Otín, C.; Viñuela, E. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology 1995, 206, 1140–1144. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.M.; García-Escudero, R.; Salas, M.L.; Andrés, G. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 2004, 78, 1313–4299. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, F.; Alcaraz, C.; Eiras, A.; Yáñez, R.J.; Rodriguez, J.M.; Alonso, C.; Rodriguez, J.F.; Escribano, J.M. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J. Virol. 1994, 68, 7244–7252. [Google Scholar] [CrossRef] [Green Version]
- Simón-Mateo, C.; Andrés, G.; Viñuela, E. Polyprotein processing in African swine fever virus: A novel gene expression strategy for a DNA virus. EMBO J. 1993, 12, 2977–2987. [Google Scholar] [CrossRef] [PubMed]
- Andrés, G.; Simón-Mateo, C.; Viñuela, E. Assembly of African swine fever virus: Role of polyprotein pp220. J. Virol. 1997, 71, 2331–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés, G.; García-Escudero, R.; Salas, M.L.; Rodríguez, J.M. Repression of African Swine Fever Virus Polyprotein pp220-Encoding Gene Leads to the Assembly of Icosahedral Core-Less Particles. J. Virol. 2002, 76, 2654–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simón-Mateo, C.; Andrés, G.; Almazán, F.; Viñuela, E. Proteolytic processing in African swine fever virus: Evidence for a new structural polyprotein, pp62. J. Virol. 1997, 71, 5799–5804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez, C.; Salas, M.L.; Rodríguez, J.M. African swine fever virus polyprotein pp62 is essential for viral core development. J. Virol. 2010, 84, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés, G.; Alejo, A.; Simón-Mateo, C.; Salas, M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001, 276, 780–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alejo, A.; Andrés, G.; Salas, M.L. African Swine Fever virus proteinase is essential for core maturation and infectivity. J. Virol. 2003, 77, 5571–5577. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, M.; Freije, J.M.; Salas, M.L.; Viñuela, E.; López-Otín, C. Structure and expression in E. coli of the gene coding for protein p10 of African swine fever virus. Arch. Virol. 1993, 130, 93–107. [Google Scholar] [CrossRef]
- Borca, M.V.; Irusta, P.M.; Kutish, G.F.; Carrillo, C.; Afonso, C.L.; Burrage, T.; Neilan, J.G.; Rock, D.L. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch. Virol. 1996, 141, 301–313. [Google Scholar] [CrossRef]
- Bonnefoy, E.; Rouvière-Yaniv, J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991, 10, 687–696. [Google Scholar] [CrossRef]
- Dey, D.; Nagaraja, V.; Ramakumar, S. Structural and evolutionary analyses reveal determinants of DNA binding specificities of nucleoid-associated proteins HU and IHF. Mol. Phylogenet. Evol. 2017, 107, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Kunisch, R.; Kamal, E.; Lewin, A. The role of the mycobacterial DNA-binding protein 1 (MDP1) from Mycobacterium bovis BCG in host cell interaction. BMC Microbiol. 2012, 12, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, N.; Agarwal, N.; Kaur, A.; Tripathi, S.; Gahlay, G.K.; Arora, A.; Mithu, V.S.; Poluri, K.M.; Kumar, D. Molecular interaction between human SUMO-I and histone like DNA binding protein of Helicobacter pylori (Hup) investigated by NMR and other biophysical tools. Int. J. Biol. Macromol. 2019, 123, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Stojkova, P.; Spidlova, P.; Stulik, J. Nucleoid-Associated Protein HU: A Lilliputian in Gene Regulation of Bacterial Virulence. Front. Cell. Infect. Microbiol. 2019, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Sun, Y.; Chai, Y.; Li, S.; Li, S.; Wang, L.; Su, J.; Yu, S.; Yan, J.; Gao, F.; et al. The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives. Proc. Natl. Acad. Sci. USA 2020, 201922523. [Google Scholar] [CrossRef]
- Schrödinger. Schrödinger Release 2020-2: Maestro; Schrödinger: New York, NY, USA, 2020. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Saper, G.; Kler, S.; Asor, R.; Oppenheim, A.; Raviv, U.; Harries, D. Effect of capsid confinement on the chromatin organization of the SV40 minichromosome. Nucleic Acids Res. 2012, 41, 1569–1580. [Google Scholar] [CrossRef] [Green Version]
- Polisky, B.; McCarthy, B. Location of histones on simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 1975, 72, 2895–2899. [Google Scholar] [CrossRef] [Green Version]
- Bensaid, A.; Almeida, A.; Drlica, K.; Rouviere-Yaniv, J. Cross-talk Between Topoisomerase I and HU inEscherichia coli. J. Mol. Biol. 1996, 256, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Komazin-Meredith, G.; Santos, W.L.; Filman, D.J.; Hogle, J.M.; Verdine, G.L.; Coen, D.M. The Positively Charged Surface of Herpes Simplex Virus UL42 Mediates DNA Binding. J. Biol. Chem. 2008, 283, 6154–6161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlin, S.; Brendel, V. Charge configurations in viral proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 9396–9400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Requião, R.D.; Carneiro, R.L.; Moreira, M.H.; Ribeiro-Alves, M.; Rossetto, S.; Palhano, F.L.; Domitrovic, T. Viruses with different genome types adopt a similar strategy to pack nucleic acids based on positively charged protein domains. Sci. Rep. 2020, 10, 5470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbano, A.C.; Ferreira, F. Functional analysis of the ASFV DNA-binding protein pA104R. Manuscript in preparation.
- Noda, H.; Munderloh, U.G.; Kurtti, T.J. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 1997, 63, 3926–3932. [Google Scholar] [CrossRef] [Green Version]
- Fast, N.M.; Kissinger, J.C.; Roos, D.S.; Keeling, P.J. Nuclear-Encoded, Plastid-Targeted Genes Suggest a Single Common Origin for Apicomplexan and Dinoflagellate Plastids. Mol. Biol. Evol. 2001, 18, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Dey, D.; Nagaraja, V.; Ramakumar, S. Phylogenetic and structural analyses reveal the determinants of DNA binding specificities of nucleoid-associated proteins HU and IHF. bioRxiv 2016, 57489. [Google Scholar] [CrossRef] [Green Version]
- Sato, S. The apicomplexan plastid and its evolution. Cell. Mol. Life Sci. 2011, 68, 1285–1296. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, C.; Mwaengo, D.M.; Macharia, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martín, E.; Kasiti, J.; Bishop, R.P. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Gabriel, C.; Beer, M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine 2014, 32, 3879–3882. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.S.; Hess, W.R. Antibody response to inactivated preparations of African swine fever virus in pigs. Am. J. Vet. Res. 1967, 28, 475–481. [Google Scholar] [PubMed]
- Bosch-Camós, L.; López, E.; Rodriguez, F. African swine fever vaccines: A promising work still in progress. Porc. Heal. Manag. 2020, 6, 17. [Google Scholar] [CrossRef]
- Monteagudo, P.L.; Lacasta, A.; López, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71ΔCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91, e01058-17. [Google Scholar] [CrossRef] [Green Version]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020, 94, e02017-19. [Google Scholar] [CrossRef]
- O’Donnell, V.; Risatti, G.R.; Holinka, L.G.; Krug, P.W.; Carlson, J.; Velazquez-Salinas, L.; Azzinaro, P.A.; Gladue, D.P.; Borca, M. V Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2017, 91, e01760-16. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Gavier-Widén, D.; Ståhl, K.; Dixon, L. No hasty solutions for African swine fever. Science 2020, 367, 622–624. [Google Scholar] [CrossRef] [PubMed]
- Escribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: Myths and facts. Virus Res. 2013, 173, 101–109. [Google Scholar] [CrossRef]
- Sunwoo, S.-Y.; Pérez-Núñez, D.; Morozov, I.; Sánchez, E.G.; Gaudreault, N.N.; Trujillo, J.D.; Mur, L.; Nogal, M.; Madden, D.; Urbaniak, K.; et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines 2019, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neilan, J.G.; Zsak, L.; Lu, Z.; Burrage, T.G.; Kutish, G.F.; Rock, D.L. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology 2004, 319, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goatley, L.C.; Reis, A.L.; Portugal, R.; Goldswain, H.; Shimmon, G.L.; Hargreaves, Z.; Ho, C.S.; Montoya, M.; Sánchez-Cordón, P.J.; Taylor, G.; et al. A pool of eight virally vectored African swine fever antigens protect pigs against fatal disease. Vaccines 2020, 8, 234. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.L.; Parkhouse, R.M.E.; Penedos, A.R.; Martins, C.; Leitão, A. Systematic analysis of longitudinal serological responses of pigs infected experimentally with African swine fever virus. J. Gen. Virol. 2007, 88, 2426–2434. [Google Scholar] [CrossRef]
- Halfmann, P.; Ebihara, H.; Marzi, A.; Hatta, Y.; Watanabe, S.; Suresh, M.; Neumann, G.; Feldmann, H.; Kawaoka, Y. Replication-deficient ebolavirus as a vaccine candidate. J. Virol. 2009, 83, 3810–3815. [Google Scholar] [CrossRef] [Green Version]
- Coelho, J.; Leitão, A. The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control. Vaccines 2020, 8, 312. [Google Scholar] [CrossRef]
- Matsuo, E.; Celma, C.C.P.; Boyce, M.; Viarouge, C.; Sailleau, C.; Dubois, E.; Bréard, E.; Thiéry, R.; Zientara, S.; Roy, P. Generation of Replication-Defective Virus-Based Vaccines That Confer Full Protection in Sheep against Virulent Bluetongue Virus Challenge. J. Virol. 2011, 85, 10213–10221. [Google Scholar] [CrossRef] [Green Version]
- Van Rijn, P.A. Prospects of Next-Generation Vaccines for Bluetongue. Front. Vet. Sci. 2019, 6, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, F.B.; Simões, M.; Frouco, G.; Martins, C.; Ferreira, F. Towards the Generation of an ASFV-pA104R DISC Mutant and a Complementary Cell Line—A Potential Methodology for the Production of a Vaccine Candidate. Vaccines 2019, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Fernández, C.; Páez, E.; Vilas, P.; Gancedo, A.G. Effect of Disodium Phosphonoacetate and Iododeoxyuridine on the Multiplication of African Swine Fever Virus in vitro. Chemotherapy 1979, 25, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Paez, E.; Garcia, F.; Gil Fernandez, C. Interferon cures cells lytically and persistently infected with African swine fever virus in vitro. Arch. Virol. 1990, 112, 115–127. [Google Scholar] [CrossRef]
- Freitas, F.B.; Frouco, G.; Martins, C.; Leitão, A.; Ferreira, F. In vitro inhibition of African swine fever virus-topoisomerase II disrupts viral replication. Antiviral Res. 2016, 134, 34–41. [Google Scholar] [CrossRef]
- Keita, D.; Heath, L.; Albina, E. Control of African swine fever virus replication by small interfering RNA targeting the A151R and VP72 genes. Antivir. Ther. 2010, 15, 727–736. [Google Scholar] [CrossRef] [Green Version]
- Galindo, I.; Hernáez, B.; Berná, J.; Fenoll, J.; Cenis, J.L.; Escribano, J.M.; Alonso, C. Comparative inhibitory activity of the stilbenes resveratrol and oxyresveratrol on African swine fever virus replication. Antiviral Res. 2011, 91, 57–63. [Google Scholar] [CrossRef]
- Docherty, J.J.; Fu, M.M.H.; Stiffler, B.S.; Limperos, R.J.; Pokabla, C.M.; DeLucia, A.L. Resveratrol inhibition of herpes simplex virus replication. Antiviral Res. 1999, 43, 145–155. [Google Scholar] [CrossRef]
- Berardi, V.; Ricci, F.; Castelli, M.; Galati, G.; Risuleo, G. Resveratrol exhibits a strong cytotoxic activity in cultured cells and has an antiviral action against polyomavirus: Potential clinical use. J. Exp. Clin. Cancer Res. 2009, 28, 96. [Google Scholar] [CrossRef] [Green Version]
- Cheltsov, A.V.; Aoyagi, M.; Aleshin, A.; Yu, E.C.-W.; Gilliland, T.; Zhai, D.; Bobkov, A.A.; Reed, J.C.; Liddington, R.C.; Abagyan, R. Vaccinia Virus Virulence Factor N1L is a Novel Promising Target for Antiviral Therapeutic Intervention. J. Med. Chem. 2010, 53, 3899–3906. [Google Scholar] [CrossRef] [Green Version]
- Campagna, M.; Rivas, C. Antiviral activity of resveratrol. Biochem. Soc. Trans. 2010, 38, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Wendorff, T.J.; Berger, J.M. Resveratrol: A novel type of topoisomerase II inhibitor. J. Biol. Chem. 2017, 292, 21011–21022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhowmick, T.; Ghosh, S.; Dixit, K.; Ganesan, V.; Ramagopal, U.A.; Dey, D.; Sarma, S.P.; Ramakumar, S.; Nagaraja, V. Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors. Nat. Commun. 2014, 5, 4124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puntumchai, A.; Kittakoop, P.; Rajviroongit, S.; Vimuttipong, S.; Likhitwitayawuid, K.; Thebtaranonth, Y. Lakoochins A and B, New Antimycobacterial Stilbene Derivatives from Artocarpus lakoocha. J. Nat. Prod. 2004, 67, 485–486. [Google Scholar] [CrossRef]
- Pavan, F.R.; De Carvalho, G.S.G.; Da Silva, A.D.; Leite, C.Q.F. Synthesis and anti-Mycobacterium tuberculosis evaluation of aza-stilbene derivatives. Sci. World J. 2011, 11, 1113–1119. [Google Scholar] [CrossRef] [Green Version]
- Suarez, M.A.; Valencia, J.; Cadena, C.C.; Maiti, R.; Datta, C.; Puerto, G.; Isaza, J.H.; San Juan, H.; Nagaraja, V.; Guzman, J.D. Diarylethenes Display In Vitro Anti-TB Activity and Are Efficient Hits Targeting the Mycobacterium tuberculosis HU Protein. Molecules 2017, 22, 1245. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbano, A.C.; Ferreira, F. Role of the DNA-Binding Protein pA104R in ASFV Genome Packaging and as a Novel Target for Vaccine and Drug Development. Vaccines 2020, 8, 585. https://doi.org/10.3390/vaccines8040585
Urbano AC, Ferreira F. Role of the DNA-Binding Protein pA104R in ASFV Genome Packaging and as a Novel Target for Vaccine and Drug Development. Vaccines. 2020; 8(4):585. https://doi.org/10.3390/vaccines8040585
Chicago/Turabian StyleUrbano, Ana Catarina, and Fernando Ferreira. 2020. "Role of the DNA-Binding Protein pA104R in ASFV Genome Packaging and as a Novel Target for Vaccine and Drug Development" Vaccines 8, no. 4: 585. https://doi.org/10.3390/vaccines8040585






