Molecular Modeling of the Shigella flexneri Serogroup 3 and 5 O-Antigens and Conformational Relationships for a Vaccine Containing Serotypes 2a and 3a
Abstract
:1. Introduction
2. Materials and Methods
2.1. φ, ψ PMF Calculations
2.2. Molecular Dynamics
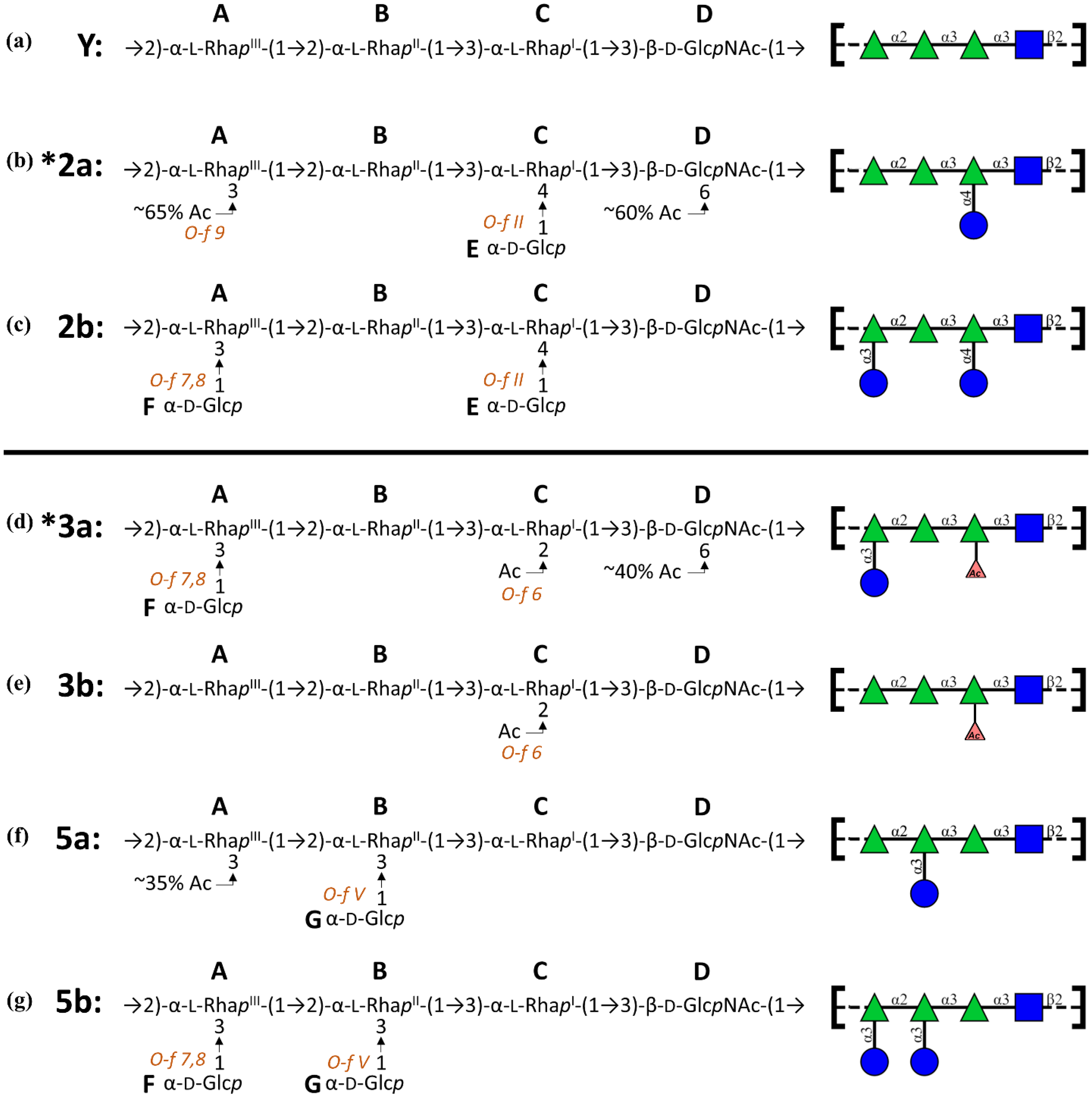
- 3a: →2)-[αDGlc(1→3)]αLRha(1→2)-αLRha(1→3)-αLRha2Ac(1→3)-βDGlcNAc-(1→
- 3b: →2)-αLRha(1→2)-αLRha(1→3)-αLRha2Ac(1→3)-βDGlcNAc-(1→
- 5a: →2)-αLRha(1→2)-[αDGlc(1→3)]αLRha(1→3)-αLRha(1→3)-βDGlcNAc-(1→
- 5b: →2)-[αDGlc(1→3)]αLRha(1→2)-[αDGlc(1→3)]αLRha(1→3)-αLRha(1→3)-βDGlcNAc-(1→
2.3. Block Averaging Analysis
2.4. Data Analysis
3. Results
3.1. Simulation Convergence
3.2. O-Ag Flexibility
3.3. O-Ag Conformations
3.4. O-Ag Glycosidic Linkage Conformations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [Green Version]
- Kotloff, K.L.; Platts-Mills, J.A.; Nasrin, D.; Roose, A.; Blackwelder, W.C.; Levine, M.M. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 2017, 35, 6783–6789. [Google Scholar] [CrossRef] [PubMed]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.; The, H.C. Recent insights into Shigella: A major contributor to the global diarrhoeal disease burden. Curr. Opin. Infect. Dis. 2018, 31, 449. [Google Scholar] [CrossRef]
- Ranjbar, R.; Farahani, A. Shigella: Antibiotic-Resistance Mechanisms And New Horizons For Treatment. Infect. Drug Resist. 2019, 12, 3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkenazi, S.; Levy, I.; Kazaronovski, V.; Samra, Z. Growing antimicrobial resistance of Shigella isolates. J. Antimicrob. Chemother. 2003, 51, 427–429. [Google Scholar] [CrossRef] [Green Version]
- Gaudreau, C.; Pilon, P.A.; Cornut, G.; Marchand-Senecal, X.; Bekal, S. Shigella flexneri with ciprofloxacin resistance and reduced azithromycin susceptibility, Canada, 2015. Emerg. Infect. Dis. 2016, 22. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Guidelines for the Control of Shigellosis, Including Epidemics due to Shigella Dysenteriae Type 1; WHO Press: Geneva, Switzerland, 2005. [Google Scholar]
- Walker, R.I. An assessment of enterotoxigenic Escherichia coli and Shigella vaccine candidates for infants and children. Vaccine 2015, 33, 954–965. [Google Scholar] [CrossRef] [Green Version]
- DeLaine, B.C.; Wu, T.; Grassel, C.L.; Shimanovich, A.; Pasetti, M.F.; Levine, M.M.; Barry, E.M. Characterization of a multicomponent live, attenuated Shigella flexneri vaccine. FEMS Pathog. Dis. 2016, 74, ftw034. [Google Scholar] [CrossRef] [Green Version]
- Perepelov, A.V.; Shekht, M.E.; Liu, B.; Shevelev, S.D.; Ledov, V.A.; Senchenkova, S.Y.N.; L’vov, V.L.; Shashkov, A.S.; Feng, L.; Aparin, P.G. Shigella flexneri O-antigens revisited: Final elucidation of the O-acetylation profiles and a survey of the O-antigen structure diversity. FEMS Immunol. Med. Mic. 2012, 66, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Knirel, Y.; Sun, Q.; Senchenkova, S.; Perepelov, A.; Shashkov, A.; Xu, J. O-Antigen modifications providing antigenic diversity of Shigella flexneri and underlying genetic mechanisms. Biochem. Mosc. 2015, 80, 901–914. [Google Scholar] [CrossRef]
- Lindberg, A.A.; Karnell, A.; Weintraub, A. The lipopolysaccharide of Shigella bacteria as a virulence factor. Rev. Infect. Dis. 1991, 13 (Suppl. 4), S279–S284. [Google Scholar] [CrossRef] [PubMed]
- Jennison, A.V.; Verma, N.K. Shigella flexneri infection: Pathogenesis and vaccine development. FEMS Microbiol. Rev. 2004, 28, 43–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, M.M.; Kotloff, K.L.; Barry, E.M.; Pasetti, M.F.; Sztein, M.B. Clinical trials of Shigella vaccines: Two steps forward and one step back on a long, hard road. Nat. Rev. Microbiol. 2007, 5, 540–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barel, L.-A.; Mulard, L.A. Classical and novel strategies to develop a Shigella glycoconjugate vaccine: From concept to efficacy in human. Hum. Vacc. Immunother. 2019, 15, 1338–1356. [Google Scholar] [CrossRef]
- Ewing, W.H.; Carpenter, K.P. Recommended designations for the subserotypes of Shigella flexneri. Int. J. Syst. Evol. Microbiol. 1966, 16, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Livio, S.; Strockbine, N.A.; Panchalingam, S.; Tennant, S.M.; Barry, E.M.; Marohn, M.E.; Antonio, M.; Hossain, A.; Mandomando, I.; Ochieng, J.B. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin. Infect. Dis. 2014, 59, 933–941. [Google Scholar] [CrossRef]
- Mani, S.; Wierzba, T.; Walker, R.I. Status of vaccine research and development for Shigella. Vaccine 2016, 34, 2887–2894. [Google Scholar] [CrossRef] [Green Version]
- Kotloff, K.L.; Riddle, M.S.; Platts-Mills, J.A.; Pavlinac, P.; Zaidi, A.K. Shigellosis. Lancet 2018, 391, 801–812. [Google Scholar] [CrossRef]
- Kuttel, M.M.; Ravenscroft, N. The Role of Molecular Modeling in Predicting Carbohydrate Antigen Conformation and Understanding Vaccine Immunogenicity. In Carbohydrate-Based Vaccines: From Concept to Clinic; ACS Publications: Washington, DC, USA, 2018; pp. 139–173. [Google Scholar]
- Bock, K.; Josephson, S.; Bundle, D.R. Lipopolysaccharide solution conformation: Antigen shape inferred from high resolution 1 H and 13 C nuclear magnetic resonance spectroscopy and hard-sphere calculations. J. Chem. Soc. Perkin Trans. 1982, 2, 59–70. [Google Scholar] [CrossRef]
- Clément, M.-J.; Imberty, A.; Phalipon, A.; Pérez, S.; Simenel, C.; Mulard, L.A.; Delepierre, M. Conformational studies of the O-specific polysaccharide of Shigella flexneri 5a and of four related synthetic pentasaccharide fragments using NMR and molecular modeling. J. Biol. Chem. 2003, 278, 47928–47936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.; Barbirz, S.; Lipowsky, R.; Santer, M. Conformational diversity of O-antigen polysaccharides of the Gram-negative bacterium Shigella flexneri serotype Y. J. Phys. Chem. B 2014, 118, 2523–2534. [Google Scholar] [CrossRef]
- Hlozek, J.; Ravenscroft, N.; Kuttel, M.M. The Effects of Glucosylation and O-Acetylation on the Conformation of Shigella Flexneri Serogroup 2 O-Antigen Vaccine Targets. J. Phys. Chem. B 2020, 124, 2806–2814. [Google Scholar] [CrossRef]
- Theillet, F.-X.; Simenel, C.; Guerreiro, C.; Phalipon, A.; Mulard, L.A.; Delepierre, M. Effects of backbone substitutions on the conformational behavior of Shigella flexneri O-antigens: Implications for vaccine strategy. Glycobiology 2011, 21, 109–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhara, D.; Kar, R.K.; Bhunia, A.; Misra, A.K. Convergent Synthesis and Conformational Analysis of the Hexasaccharide. Repeating Unit of the O-Antigen of Shigella flexneri Serotype 1d. Eur. J. Org. Chem. 2014, 4577–4584. [Google Scholar] [CrossRef]
- Von Seidlein, L.; Kim, D.R.; Ali, M.; Lee, H.; Wang, X.; Thiem, V.D.; Canh, D.G.; Chaicumpa, W.; Agtini, M.D.; Hossain, A. A multicentre study of Shigella diarrhoea in six Asian countries: Disease burden, clinical manifestations, and microbiology. PLoS Med. 2006, 3, e353. [Google Scholar] [CrossRef]
- Chompook, P.; Samosornsuk, S.; Von Seidlein, L.; Jitsanguansuk, S.; Sirima, N.; Sudjai, S.; Mangjit, P.; Kim, D.R.; Wheeler, J.G.; Todd, J. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull. World Health Organ. 2005, 83, 739–746. [Google Scholar] [PubMed]
- Boutet, J.; Blasco, P.; Guerreiro, C.; Thouron, F.; Dartevelle, S.; Nato, F.; Cañada, F.J.; Ardá, A.; Phalipon, A.; Jiménez-Barbero, J. Detailed Investigation of the Immunodominant Role of O-Antigen Stoichiometric O-Acetylation as Revealed by Chemical Synthesis, Immunochemistry, Solution Conformation and STD-NMR Spectroscopy for Shigella flexneri 3a. Chem. Eur. J. 2016, 22, 10892–10911. [Google Scholar] [CrossRef]
- Carlin, N.; Wehler, T.; Lindberg, A. Shigella flexneri O-antigen epitopes: Chemical and immunochemical analyses reveal that epitopes of type III and group 6 antigens are identical. Infect. Immun. 1986, 53, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lan, R.; Knirel, Y.A.; Luo, X.; Sof’ya, N.S.; Shashkov, A.S.; Xu, J.; Sun, Q. Serological identification and prevalence of a novel O-antigen epitope linked to 3-and 4-O-acetylated rhamnose III of lipopolysaccharide in Shigella flexneri. J. Clin. Microbiol. 2014, 52, 2033–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onodera, N.T.; Ryu, J.; Durbic, T.; Nislow, C.; Archibald, J.M.; Rohde, J.R. Genome sequence of Shigella flexneri serotype 5a strain M90T Sm. J. Bacteriol. Res. 2012. [Google Scholar] [CrossRef] [Green Version]
- Varki, A.; Freeze, H.H.; Manzi, A.E. Overview of glycoconjugate analysis. Curr. Protoc. Protein Sci. 2009, 57. [Google Scholar] [CrossRef]
- Van De Verg, L.L.; Bendiuk, N.O.; Kotloff, K.; Marsh, M.M.; Ruckert, J.L.; Puryear, J.L.; Taylor, D.N.; Hartman, A.B. Cross-reactivity of Shigella flexneri serotype 2a O antigen antibodies following immunization or infection. Vaccine 1996, 14, 1062–1068. [Google Scholar] [CrossRef]
- Noriega, F.R.; Liao, F.M.; Maneval, D.R.; Ren, S.; Formal, S.B.; Levine, M.M. Strategy for cross-protection among Shigella flexneri serotypes. Infect. Immun. 1999, 67, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Hlozek, J.; Kuttel, M.M.; Ravenscroft, N. Conformations of Neisseria meningitidis serogroup A and X polysaccharides: The effects of chain length and O-acetylation. Carbohydr. Res. 2018, 465, 44–51. [Google Scholar] [CrossRef]
- Hlozek, J.; Ravenscroft, N.; Kuttel, M.M. Modeling the conformations of Neisseria meningitidis serogroup a CPS and a carba-analogue: Implications for vaccine development. Carbohydr. Res. 2019, 486, 107838. [Google Scholar] [CrossRef] [PubMed]
- Kuttel, M.; Ravenscroft, N.; Foschiatti, M.; Cescutti, P.; Rizzo, R. Conformational properties of two exopolysaccharides produced by Inquilinus limosus, a cystic fibrosis lung pathogen. Carbohydr. Res. 2012, 350, 40–48. [Google Scholar] [CrossRef]
- Kuttel, M.M.; Timol, Z.; Ravenscroft, N. Cross-protection in Neisseria meningitidis serogroups Y and W polysaccharides: A comparative conformational analysis. Carbohydr. Res. 2017, 446, 40–47. [Google Scholar] [CrossRef]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Hardy, D.J.; Trabuco, L.G.; Schulten, K. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 2007, 28, 2618–2640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guvench, O.; Greene, S.N.; Kamath, G.; Brady, J.W.; Venable, R.M.; Pastor, R.W.; Mackerell, A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008, 29, 2543–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guvench, O.; Hatcher, E.; Venable, R.M.; Pastor, R.W.; MacKerell, A.D., Jr. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009, 5, 2353–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Kuttel, M.M.; Ståhle, J.; Widmalm, G. CarbBuilder: Software for building molecular models of complex oligo-and polysaccharide structures. J. Comput. Chem. 2016, 37, 2098–2105. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Flyvbjerg, H.; Petersen, H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989, 91, 461–466. [Google Scholar] [CrossRef]
- Grossfield, A.; Zuckerman, D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009, 5, 23–48. [Google Scholar]
- Cross, S.; Kuttel, M.M.; Stone, J.E.; Gain, J.E. Visualisation of cyclic and multi-branched molecules with VMD. J. Mol. Graph. Model. 2009, 28, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, A.; Valverde, P.; Ardá, A.; Jiménez-Barbero, J. Glycan structures and their interactions with proteins. A NMR view. Curr. Opin. Struct. Biol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Dharmasena, M.N.; Osorio, M.; Takeda, K.; Stibitz, S.; Kopecko, D.J. Stable chromosomal expression of Shigella flexneri 2a and 3a O-antigens in the live Salmonella oral vaccine vector Ty21a. Clin. Vaccine Immunol. 2017, 24, e00181-17. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Smith, M.A.; Benjamin, W.H.; Kaminski, R.W.; Wenzel, H.; Nahm, M.H. Monoclonal antibodies to Shigella lipopolysaccharide are useful for vaccine production. Clin. Vaccine Immunol. 2016, 23, 681–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahm, M.H.; Yu, J.; Weerts, H.P.; Wenzel, H.; Tamilselvi, C.S.; Chandrasekaran, L.; Pasetti, M.F.; Mani, S.; Kaminski, R.W. Development, interlaboratory evaluations, and application of a simple, high-throughput Shigella serum bactericidal assay. mSphere 2018, 3, e00146-18. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hlozek, J.; Owen, S.; Ravenscroft, N.; Kuttel, M.M. Molecular Modeling of the Shigella flexneri Serogroup 3 and 5 O-Antigens and Conformational Relationships for a Vaccine Containing Serotypes 2a and 3a. Vaccines 2020, 8, 643. https://doi.org/10.3390/vaccines8040643
Hlozek J, Owen S, Ravenscroft N, Kuttel MM. Molecular Modeling of the Shigella flexneri Serogroup 3 and 5 O-Antigens and Conformational Relationships for a Vaccine Containing Serotypes 2a and 3a. Vaccines. 2020; 8(4):643. https://doi.org/10.3390/vaccines8040643
Chicago/Turabian StyleHlozek, Jason, Sara Owen, Neil Ravenscroft, and Michelle M. Kuttel. 2020. "Molecular Modeling of the Shigella flexneri Serogroup 3 and 5 O-Antigens and Conformational Relationships for a Vaccine Containing Serotypes 2a and 3a" Vaccines 8, no. 4: 643. https://doi.org/10.3390/vaccines8040643
APA StyleHlozek, J., Owen, S., Ravenscroft, N., & Kuttel, M. M. (2020). Molecular Modeling of the Shigella flexneri Serogroup 3 and 5 O-Antigens and Conformational Relationships for a Vaccine Containing Serotypes 2a and 3a. Vaccines, 8(4), 643. https://doi.org/10.3390/vaccines8040643





