Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
2.2. SARS-COV-2 Virus
2.3. Preparation of Cell-Free Supernatant (CFS) from Lactobacillus plantarum Probio-88 (P88-CFS)
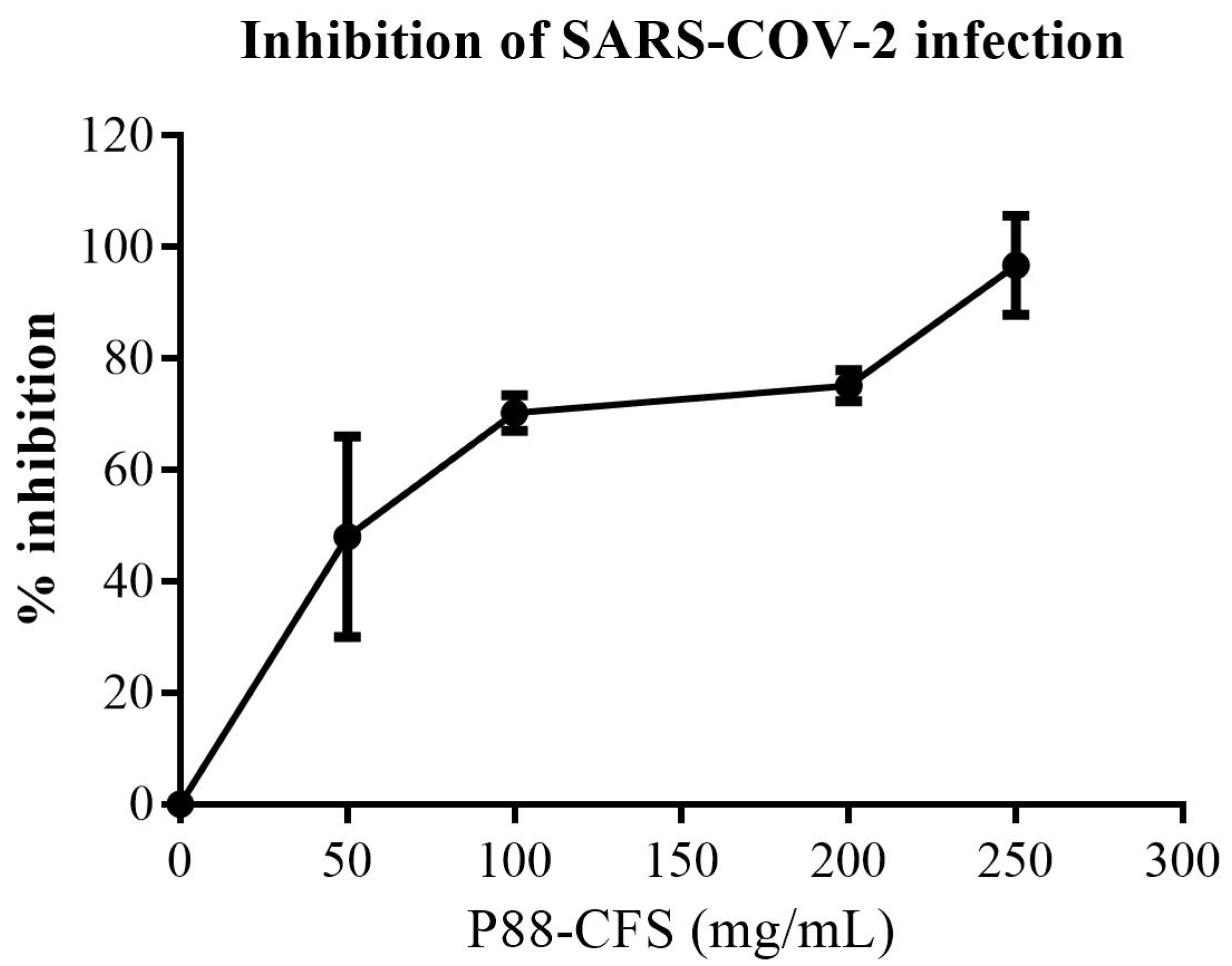
2.4. In Vitro Inhibition of SARS-COV-2 Replication
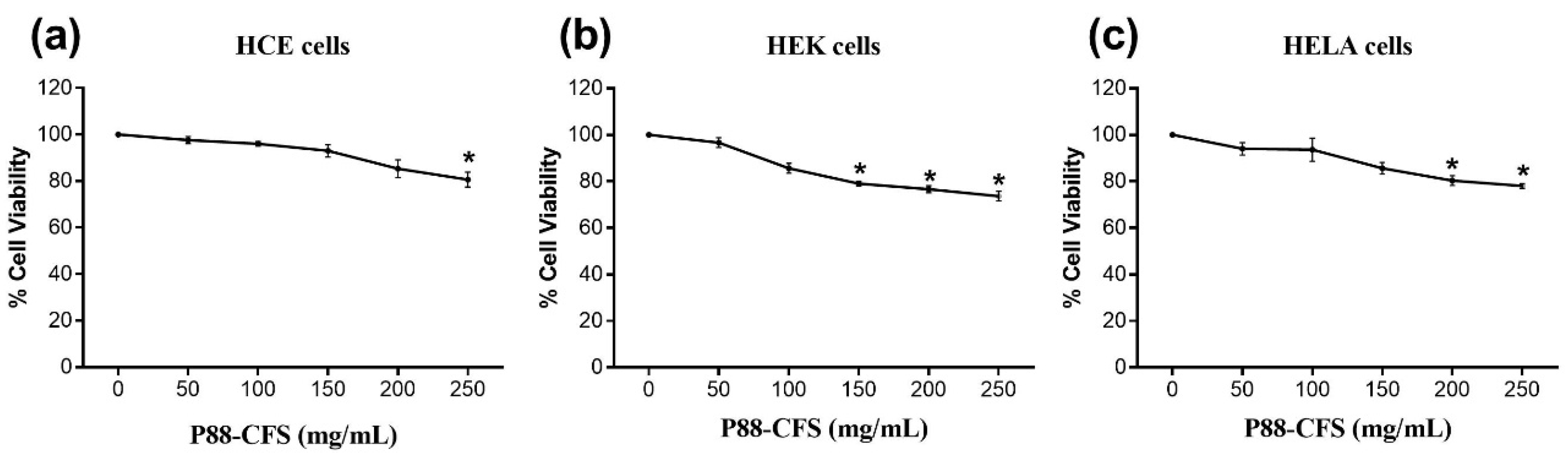
2.5. Cell-Viability Assay
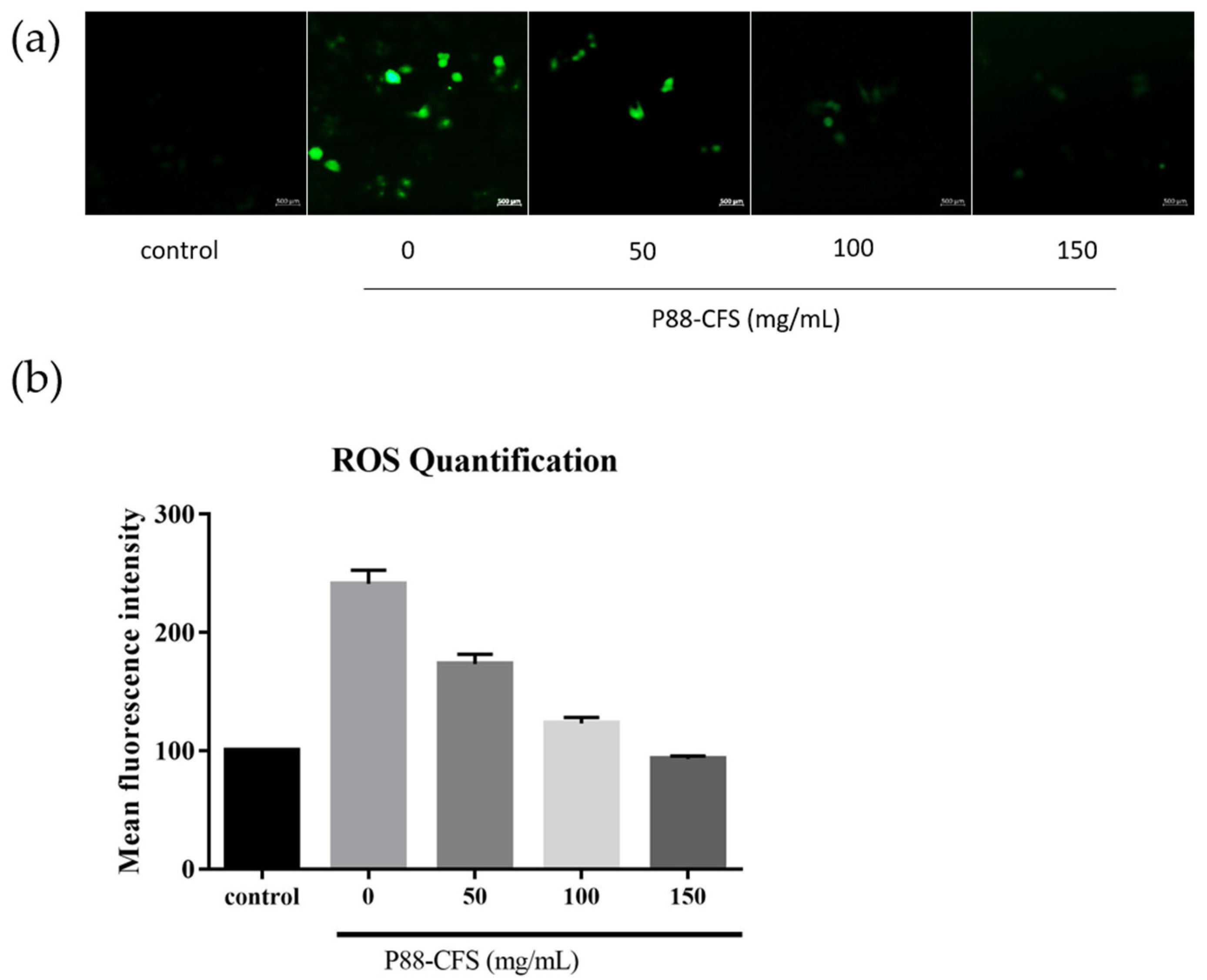
2.6. Determination of Intracellular ROS
2.7. Immunocytochemistry of Hela Cells
2.8. Quantitative Real-Time PCR
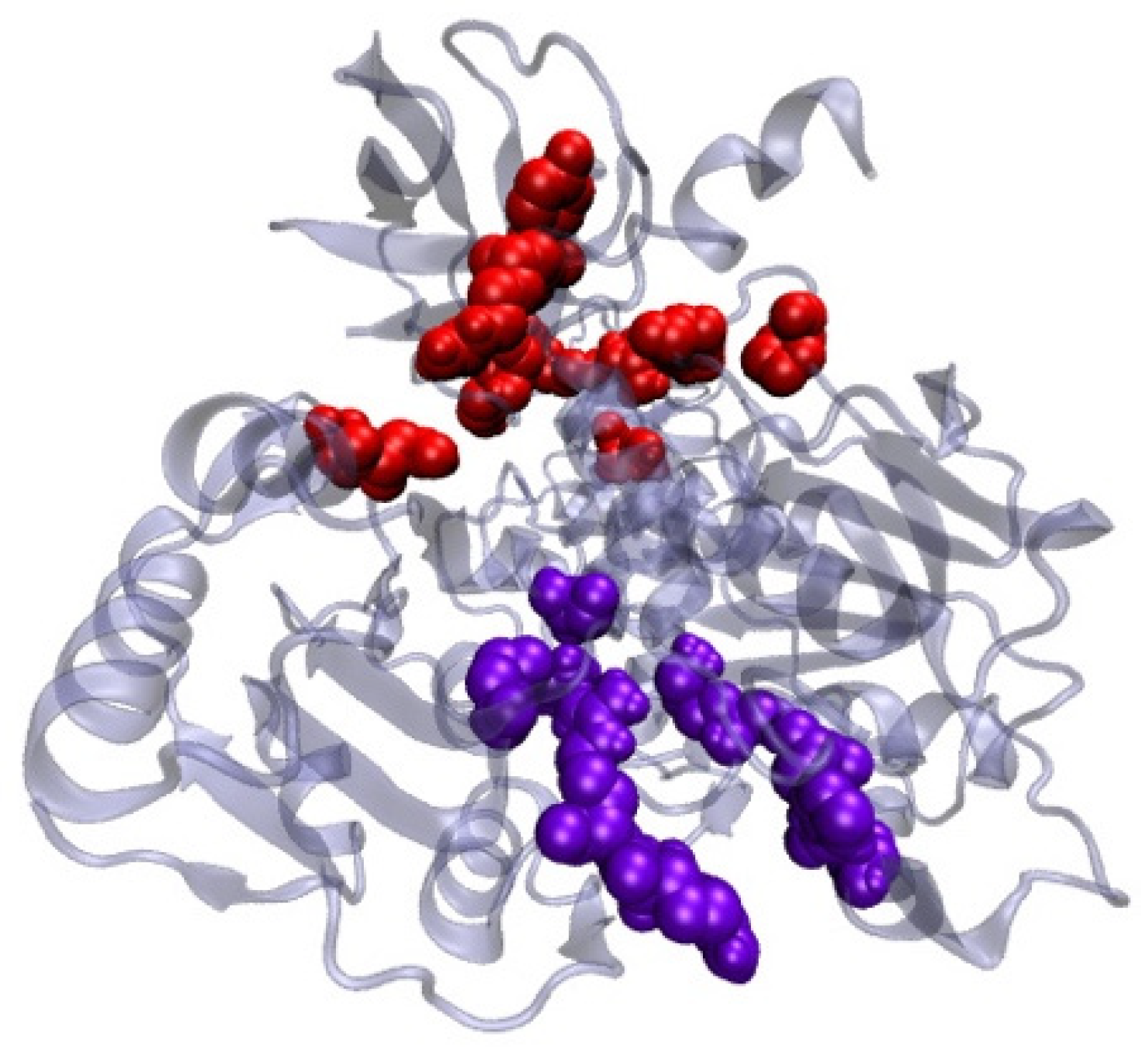
2.9. Antiviral Activity of Lactobacillus plantarum Probio-88 Using In Silico Molecular Docking
Modelling of Plantaricin E and Plantaricin F
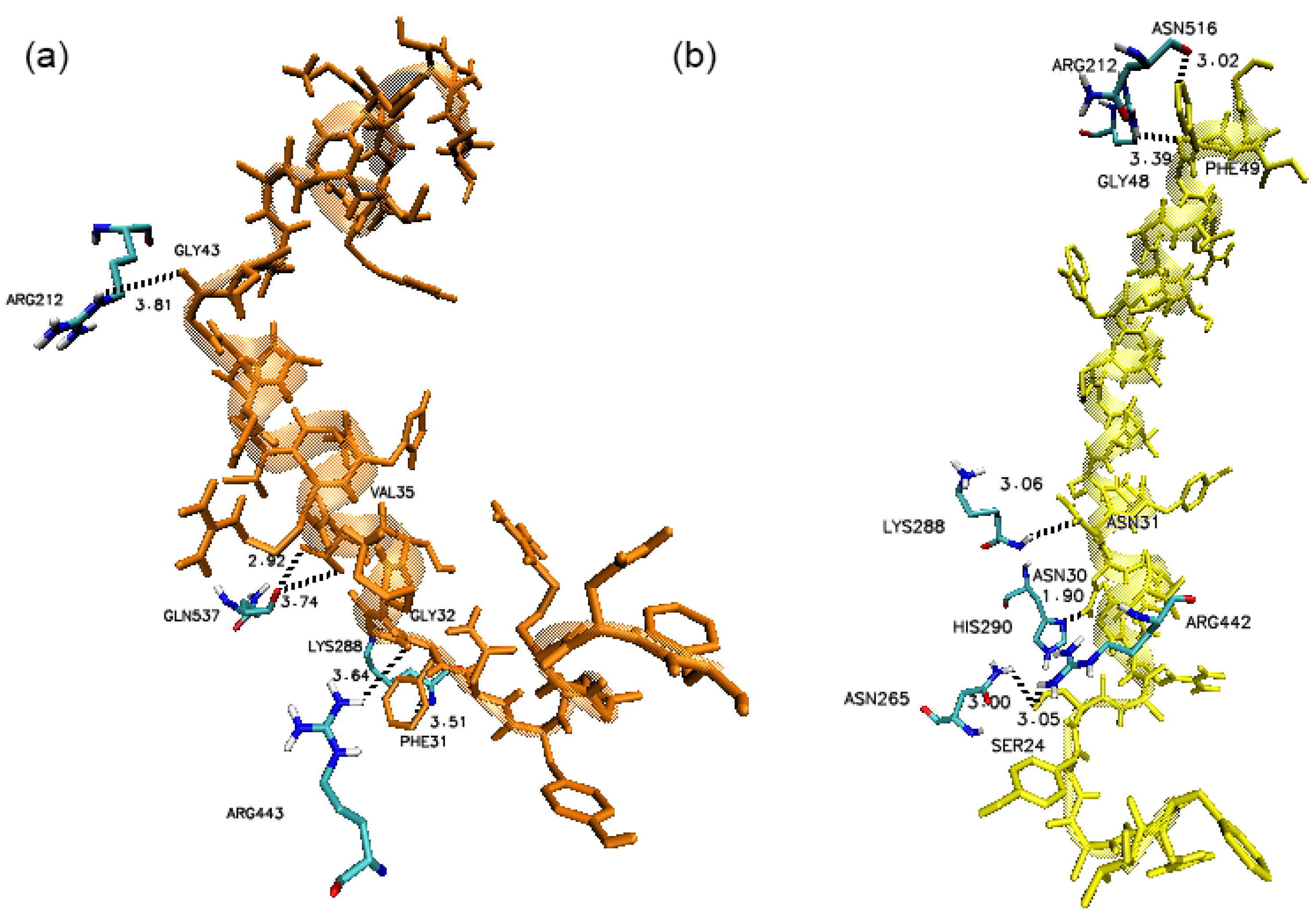
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-COV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Matheson, N.J.; Lehner, P.J. How does SARS-COV-2 cause COVID-19? Science 2020, 369, 510–511. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, W.; Xie, J.; Hou, Y.; You, C. Cytokine storm induced by SARS-COV-2. Clin. Chim. Acta 2020, 509, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Angurana, S.K.; Bansal, A.; Muralidharan, J. Evaluation of Effect of Probiotics on Cytokine Levels in Critically Ill Children With Severe Sepsis: A Double-Blind, Placebo-Controlled Trial. Crit. Care Med. 2018, 46, 1656–1664. [Google Scholar] [CrossRef]
- Custodero, C.; Mankowski, R.; Lee, S.; Chen, Z.; Wu, S.; Manini, T.; Echeverri, J.H.; Sabbà, C.; Beavers, D.; Cauley, J.; et al. Evidence-based nutritional and pharmacological interventions targeting chronic low-grade inflammation in middle-age and older adults: A systematic review and meta-analysis. Ageing Res. Rev. 2018, 46, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Lehtoranta, L.; Latvala, S.; Lehtinen, M.J. Role of Probiotics in Stimulating the Immune System in Viral Respiratory Tract Infections: A Narrative Review. Nutrients 2020, 12, 3163. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Sun, Y.-Z.; Wang, A.; Zhou, Z. Probiotics as Means of Diseases Control in Aquaculture, a Review of Current Knowledge and Future Perspectives. Front. Microbiol. 2018, 9, 2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Seo, B.J.; Mun, M.R.; Kim, C.-J.; Lee, I.; Kim, H.; Park, Y.-H. Putative probiotic Lactobacillus spfrom porcine gastrointestinal tract inhibit transmissible gastroenteritis coronavirus and enteric bacterial pathogens. Trop. Anim. Health Prod. 2010, 42, 1855–1860. [Google Scholar] [PubMed] [Green Version]
- d’Ettorre, G.; Ceccarelli, G.; Marazzato, M.; Campagna, G.; Pinacchio, C.; Alessandri, F.; Ruberto, F.; Rossi, G.; Celani, L.; Scagnolari, C.; et al. Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 2020, 7, 389. [Google Scholar] [CrossRef]
- Al Kassaa, I. Antiviral Probiotics: A New Concept in Medical Sciences. In New Insights on Antiviral Probiotics: From Research to Applications; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–46. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Beer, T.A.P.D.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; van Dijk, M.; de Vries, S.; Bonvin, A.M. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [Green Version]
- White, M.A.; Lin, W.; Cheng, X. Discovery of COVID-19 Inhibitors Targeting the SARS-COV-2 Nsp13 Helicase. J. Phys. Chem. Lett. 2020, 11, 9144–9151. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Rodrigues, J.; Kastritis, P.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Newman, J.A.; Douangamath, A.; Yazdani, S.; Yosaatmadja, Y.; Aimon, A.; Brandão-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D. Structure, mechanism and crystallographic fragment screening of the SARS-COV-2 NSP13 helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [Green Version]
- Nainu, F.; Shiratsuchi, A.; Nakanishi, Y. Induction of Apoptosis and Subsequent Phagocytosis of Virus-Infected Cells As an Antiviral Mechanism. Front. Immunol. 2017, 8, 1220. [Google Scholar] [CrossRef]
- Schwarz, K.B. Oxidative stress during viral infection: A review. Free Radic. Biol. Med. 1996, 21, 641–649. [Google Scholar] [CrossRef]
- Lim, J.-Y.; Oh, E.; Kim, Y.; Jung, W.-W.; Kim, H.-S.; Lee, J.; Sul, D. Enhanced oxidative damage to DNA, lipids, and proteins and levels of some antioxidant enzymes, cytokines, and heat shock proteins in patients infected with influenza H1N1 virus. Acta Virol. 2014, 58, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.-Y.; Chang, S.C.; Wu, H.-Y.; Yu, T.-C.; Wei, W.-C.; Lin, S.; Chien, C.-L.; Chang, M.-F. Upregulation of the Chemokine (C-C Motif) Ligand 2 via a Severe Acute Respiratory Syndrome Coronavirus Spike-ACE2 Signaling Pathway. J. Virol. 2010, 84, 7703–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghasemnejad-Berenji, M.; Pashapour, S. SARS-COV-2 and the Possible Role of Raf/MEK/ERK Pathway in Viral Survival: Is This a Potential Therapeutic Strategy for COVID-19? Pharmacology 2021, 106, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Klemm, C.; Bruchhagen, C.; Van Krüchten, A.; Niemann, S.; Löffler, B.; Peters, G.; Ludwig, S.; Ehrhardt, C. Mitogen-activated protein kinases (MAPKs) regulate IL-6 over-production during concomitant influenza virus and Staphylococcus aureus infection. Sci. Rep. 2017, 7, 42473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-COV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Xia, J.; Guo, Q.; Yen, H.-L.; et al. SARS-COV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Altayb, H.N.; Al-Abbasi, F.A.; Al-Malki, A.L.; Kamal, M.A.; Kumar, V. Antiviral effects of probiotic metabolites on COVID-19. J. Biomol. Struct. Dyn. 2020, 39, 4175–4184. [Google Scholar] [CrossRef]
- Kim, S.W.; Kang, S.I.; Shin, D.H.; Oh, S.Y.; Lee, C.W.; Yang, Y.; Son, Y.K.; Yang, H.-S.; Lee, B.-H.; An, H.-J.; et al. Potential of Cell-Free Supernatant from Lactobacillus plantarum NIBR97, Including Novel Bacteriocins, as a Natural Alternative to Chemical Disinfectants. Pharmaceuticals 2020, 13, 266. [Google Scholar] [CrossRef]
- Zrelli, S.; Amairia, S.; Zrelli, M. Respiratory syndrome coronavirus-2 response: Microbiota as lactobacilli could make the difference. J. Med. Virol. 2021, 93, 3288–3293. [Google Scholar] [CrossRef]
- Salman, J.A.S.; Mahmood, N.N.; Abdulsattar, B.O.; Abid, H.A. The effectiveness of probiotics against viral infections: A rapid review with focus on SARS-COV-2 infection. Open Access Maced. J. Med. Sci. 2020, 8, 496–508. [Google Scholar] [CrossRef]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and Their Potential Applications in Early Life Nutrition and Beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef] [Green Version]
- AL Kassaa, I.; Hober, D.; Hamze, M.; Chihib, N.E.; Drider, D. Antiviral Potential of Lactic Acid Bacteria and Their Bacteriocins. Probiotics Antimicrob. Proteins 2014, 6, 177–185. [Google Scholar] [CrossRef]
- Dreyer, L.; Smith, C.; Deane, S.M.; Dicks, L.M.T.; Van Staden, A.D. Migration of Bacteriocins Across Gastrointestinal Epithelial and Vascular Endothelial Cells, as Determined Using In Vitro Simulations. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Singh, K.; Rao, A. Probiotics: A potential immunomodulator in COVID-19 infection management. Nutr. Res. 2021, 87, 1–12. [Google Scholar] [CrossRef]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, J.; Anto, J.M.; Czarlewski, W.; Haahtela, T.; Fonseca, S.C.; Iaccarino, G.; Blain, H.; Vidal, A.; Sheikh, A.; Akdis, C.A.; et al. Cabbage and fermented vegetables: From death rate heterogeneity in countries to candidates for mitigation strategies of severe COVID-19. Allergy 2021, 76, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Li, C.; Vizoso-Pinto, M.G.; Sacur, J.; Ren, L.; Kitazawa, H. Lactiplantibacillus plantarum as a Potential Adjuvant and Delivery System for the Development of SARS-COV-2 Oral Vaccines. Microorganisms 2021, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.K.; Dicks, L.M.T.; Popov, I.V.; Karaseva, A.; Ermakov, A.M.; Suvorov, A.; Tagg, J.R.; Weeks, R.; Chikindas, M.L. Probiotics at War Against Viruses: What Is Missing From the Picture? Front. Microbiol. 2020, 11, 1877. [Google Scholar] [CrossRef]
- Tsai, C.-J.; Lin, S.L.; Wolfson, H.J.; Nussinov, R. Studies of protein-protein interfaces: A statistical analysis of the hydrophobic effect. Protein Sci. 1997, 6, 53–64. [Google Scholar] [CrossRef]
- Frick, D.N.; Lam, A.M. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006, 12, 1315–1338. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-COV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Gong, W. Will Mutations in the Spike Protein of SARS-COV-2 Lead to the Failure of COVID-19 Vaccines? J. Korean Med. Sci. 2021, 36, e124. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S.; Nabavi, S.F.; Banach, M.; Berindan-Neagoe, I.; Sarkar, K.; Sil, P.C.; Nabavi, S.M. Should We Try SARS-COV-2 Helicase Inhibitors for COVID-19 Therapy? Arch. Med. Res. 2020, 51, 733–735. [Google Scholar] [CrossRef] [PubMed]
- WHO. SARS-COV-2 Variants; WHO: Geneva, Switzerland, 2020; Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 18 August 2021).




| Target | Primer Directions | Sequences (5→3) |
|---|---|---|
| GAPDH | Forward Reverse | CAC CAC CAA CTG CTT AGC AC CCC TGT TGC TGT AGC CAA AT |
| IFN α | Forward Reverse | GAT GGC AAC CAG TTC CAG AAG AAA GAG GTT GAA GAT CTG CTG GAT |
| IFN β | Forward Reverse | CTC CAC TAC AGC TCT TTC CAT GTC AAA GTT CAT CCT GTC CTT |
| IL-6 | Forward Reverse | AAC TCC TTC TCC AGA AGC GCC GTG GGG CGG CTA CAT CTT T |
| Region of Plot | Plantaricin E | Plantaricin F | ||||||
|---|---|---|---|---|---|---|---|---|
| Built Model | Template (PDB ID: 2JUI) | Built Model | Template (PDB ID: 2RLW) | |||||
| No. of Residues | % | No. of Residues | % | No. of Residues | % | No. of Residues | % | |
| Residue in most favoured region | 21 | 84.0 | 21 | 84.0 | 26 | 92.9 | 25 | 89.3 |
| Residue in additionally allowed region | 4 | 16.0 | 4 | 16.0 | 1 | 3.6 | 2 | 7.1 |
| Residue in generously allowed regions | 0 | 0.0 | 0 | 0.0 | 1 | 3.6 | 1 | 3.6 |
| Residue in disallowed region | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Total | 25 | 100 | 25 | 100 | 28 | 100 | 34 | 100 |
| Analysis | Types of Plantaricin | |
|---|---|---|
| E | F | |
| Binding Interaction | ||
| ∆G, (kcal/mol) | −17.4 | −15.6 |
| Kd (M) at 37 °C | 5.8 × 10−13 | 1.0 × 10−11 |
| Number of interactions | ||
| Hydrogen Bonding | 15 | 12 |
| Hydrophobics | 16 | 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rather, I.A.; Choi, S.-B.; Kamli, M.R.; Hakeem, K.R.; Sabir, J.S.M.; Park, Y.-H.; Hor, Y.-Y. Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2. Vaccines 2021, 9, 1067. https://doi.org/10.3390/vaccines9101067
Rather IA, Choi S-B, Kamli MR, Hakeem KR, Sabir JSM, Park Y-H, Hor Y-Y. Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2. Vaccines. 2021; 9(10):1067. https://doi.org/10.3390/vaccines9101067
Chicago/Turabian StyleRather, Irfan A., Sy-Bing Choi, Majid Rasool Kamli, Khalid Rehman Hakeem, Jamal S. M. Sabir, Yong-Ha Park, and Yan-Yan Hor. 2021. "Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2" Vaccines 9, no. 10: 1067. https://doi.org/10.3390/vaccines9101067
APA StyleRather, I. A., Choi, S.-B., Kamli, M. R., Hakeem, K. R., Sabir, J. S. M., Park, Y.-H., & Hor, Y.-Y. (2021). Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2. Vaccines, 9(10), 1067. https://doi.org/10.3390/vaccines9101067








