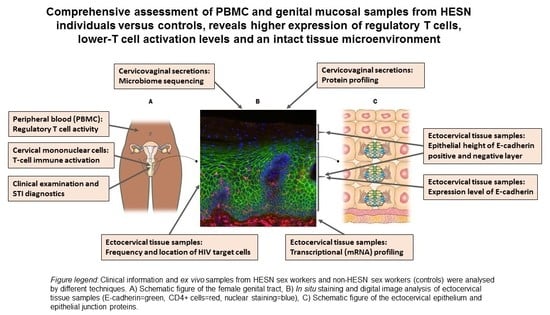
HIV-Exposed Seronegative Sex Workers Express Low T-Cell Activation and an Intact Ectocervical Tissue Microenvironment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Sample Collection
2.3. Phenotyping of Peripheral Blood Mononuclear Cells and Cervical Mononuclear Cells
2.4. Protein Profiling Using a Bead-Based Affinity Assay
2.5. Cytokine Measurement
2.6. 16S Ribosomal RNA (rRNA) Gene Sequencing to Determine the Cervicovaginal Microbiome Composition
2.7. In Situ Fluorescence Staining and Image Analysis
2.8. RNA Sequencing (RNA-seq) Analysis
3. Results
3.1. Study Population
3.2. Higher Relative Proportion of Regulatory T Cells (Treg) in Blood in HESN versus NN Women
3.3. Lower Levels of Activated Cervicovaginal CD4+ and CD8+ T Cells in HESN versus NN Women
3.4. Comparable Protein Levels in CVL between HESN and NN Women
3.5. Comparable Cervicovaginal Microbiome Composition in HESN and NN Women
3.6. Comparable Ectocervical Epithelial Thickness and HIV Receptor Expression in HESN and NN Women
3.7. Comparable Transcriptional Activity in Ectocervical Tissue of HESN and NN Women
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joint United Nations Programme on HIV/AIDS (UNAIDS) AIDS data 2019. Available online: https://www.unaids.org/en/resources/documents/2019/2019-UNAIDS-data (accessed on 15 January 2021).
- Card, C.M.; McLaren, P.J.; Wachihi, C.; Kimani, J.; Plummer, F.A.; Fowke, K.R. Decreased immune activation in resistance to HIV-1 infection is associated with an elevated frequency of CD4+CD25+FOXP3+ Regulatory T Cells. J. Infect. Dis. 2009, 199, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- McLaren, P.J.; Blake Ball, T.; Wachihi, C.; Jaoko, W.; Kelvin, D.J.; Danesh, A.; Kimani, J.; Plummer, F.A.; Fowke, K.R. HIV-exposed seronegative commercial sex workers show a quiescent phenotype in the CD4+ T cell compartment and reduced expression of HIV-dependent host factors. J. Infect. Dis. 2010, 202, 339–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flórez-Álvarez, L.; Blanquiceth, Y.; Ramírez, K.; Ossa-Giraldo, A.C.; Velilla, P.A.; Hernandez, J.C.; Zapata, W. NK Cell Activity and CD57+/NKG2Chigh Phenotype Are Increased in Men Who Have Sex With Men at High Risk for HIV. Front. Immunol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.Q.; Vendrame, E.; Ferreira, A.M.; Seiler, C.; Ranganath, T.; Alary, M.; Labbé, A.C.; Guédou, F.; Poudrier, J.; Holmes, S.; et al. Natural killer cell phenotype is altered in HIV-exposed seronegative women. PLoS ONE 2020, 15, e0238347. [Google Scholar] [CrossRef] [PubMed]
- Serna-Ortega, P.A.; Aguilar-Jimenez, W.; Florez-Álvarez, L.; Trabattoni, D.; Rugeles, M.T.; Biasin, M. IL-21 is associated with natural resistance to HIV-1 infection in a Colombian HIV exposed seronegative cohort. Microbes Infect. 2020, 22, 371–374. [Google Scholar] [CrossRef]
- Khamassi, M.; Xu, L.; Rey, J.; Duchemin, M.; Bouceba, T.; Tuffery, P.; Tudor, D.; Bomsel, M. The CH1a domain of mucosal gp41 IgA contributes to antibody specificity and antiviral functions in HIV-1 highly exposed Sero-Negative individuals. PLoS Pathog. 2020, 16, e1009103. [Google Scholar] [CrossRef]
- Burgener, A.; Rahman, S.; Ahmad, R.; Lajoie, J.; Ramdahin, S.; Mesa, C.; Brunet, S.; Wachihi, C.; Kimani, J.; Fowke, K.; et al. Comprehensive proteomic study identifies serpin and cystatin antiproteases as novel correlates of HIV-1 resistance in the cervicovaginal mucosa of female Sex workers. J. Proteome Res. 2011, 10, 5139–5149. [Google Scholar] [CrossRef]
- Lajoie, J.; Juno, J.; Burgener, A.; Rahman, S.; Mogk, K.; Wachihi, C.; Mwanjewe, J.; Plummer, F.A.; Kimani, J.; Ball, T.B.; et al. A distinct cytokine and chemokine profile at the genital mucosa is associated with HIV-1 protection among HIV-exposed seronegative commercial sex workers. Mucosal Immunol. 2012, 5, 277–287. [Google Scholar] [CrossRef]
- Pattacini, L.; Murnane, P.M.; Kahle, E.M.; Bolton, M.J.; Delrow, J.J.; Lingappa, J.R.; Katabira, E.; Donnell, D.; McElrath, M.J.; Baeten, J.M.; et al. Differential regulatory T cell activity in hiv type 1-exposed seronegative individuals. AIDS Res. Hum. Retroviruses 2013, 29, 1321–1329. [Google Scholar] [CrossRef] [Green Version]
- Fourcade, L.; Sabourin-Poirier, C.; Perraud, V.; Faucher, M.C.; Chagnon-Choquet, J.; Labbé, A.C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Natural immunity to hiv is associated with low BLyS/BAFF levels and low frequencies of innate marginal zone like CD1c+ B-cells in the genital tract. PLoS Pathog. 2019, 15, e1007840. [Google Scholar] [CrossRef] [Green Version]
- Girard, A.; Rallón, N.; Benito, J.M.; Jospin, F.; Rodriguez, C.; Chanut, B.; Benjelloun, F.; Del Romero, J.; Verrier, B.; Lucht, F.; et al. A high mucosal blocking score is associated with HIV protection. Aids 2019, 33, 411–423. [Google Scholar] [CrossRef]
- Fenizia, C.; Saulle, I.; Clerici, M.; Biasin, M. Genetic and epigenetic regulation of natural resistance to HIV-1 infection: New approaches to unveil the HESN secret. Expert Rev. Clin. Immunol. 2020, 16, 429–445. [Google Scholar] [CrossRef]
- Saulle, I.; Ibba, S.V.; Vittori, C.; Fenizia, C.; Mercurio, V.; Vichi, F.; Caputo, S.L.; Trabattoni, D.; Clerici, M.; Biasin, M. Sterol metabolism modulates susceptibility to HIV-1 Infection. AIDS 2020, 34, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hu, X.; Li, W.; Liu, J.; Liang, B.; Chen, H.; Huang, J.; Zang, N.; Ning, C.; Liao, Y.; et al. Enhanced Signaling Through the TLR9 Pathway Is Associated With Resistance to HIV-1 Infection in Chinese HIV-1–Exposed Seronegative Individuals. Front. Immunol. 2020, 11, 1050. [Google Scholar] [CrossRef] [PubMed]
- Fowke, K.R.; Nagelkerke, N.J.D.; Kimani, J.; Simonsen, J.N.; Anzala, A.O.; Bwayo, J.J.; MacDonald, K.S.; Ngugi, E.N.; Plummer, F.A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 1996, 348, 1347–1351. [Google Scholar] [CrossRef]
- Hasselrot, K.; Cheruiyot, J.; Kimani, J.; Ball, T.B.; Kaul, R.; Hirbod, T. Feasibility and Safety of Cervical Biopsy Sampling for Mucosal Immune Studies in Female Sex Workers from Nairobi, Kenya. PLoS ONE 2012, 7, e47570. [Google Scholar] [CrossRef] [Green Version]
- Lajoie, J.; Boily-Larouche, G.; Doering, K.; Cheruiyot, J.; Oyugi, J.; Broliden, K.; Kimani, J.; Fowke, K.R. Improving Adherence to Post-Cervical Biopsy Sexual Abstinence in Kenyan Female Sex Workers. Am. J. Reprod. Immunol. 2016, 76, 82–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juno, J.A.; Boily-Larouche, G.; Lajoie, J.; Fowke, K.R. Collection, isolation, and flow cytometric analysis of human endocervical samples. J. Vis. Exp. 2014, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Månberg, A.; Bradley, F.; Qundos, U.; Guthrie, B.L.; Birse, K.; Noël-Romas, L.; Lindskog, C.; Bosire, R.; Kiarie, J.; Farquhar, C.; et al. A high-throughput bead-based affinity assay enables analysis of genital protein signatures in women at risk of HIV Infection. Mol. Cell. Proteom. 2019, 18, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.B.; Burgener, A.; Birse, K.; Romas, L.; Dunphy, L.J.; Shahabi, K.; Abou, M.; Westmacott, G.R.; Mccorrister, S.; Kwatampora, J.; et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016, 9, 194–205. [Google Scholar] [CrossRef]
- Bradley, F.; Birse, K.; Hasselrot, K.; Noël-Romas, L.; Introini, A.; Wefer, H.; Seifert, M.; Engstrand, L.; Tjernlund, A.; Broliden, K.; et al. The vaginal microbiome amplifies sex hormone-associated cyclic changes in cervicovaginal inflammation and epithelial barrier disruption. Am. J. Reprod. Immunol. 2018, 80, 1–13. [Google Scholar] [CrossRef]
- Birse, K.D.; Romas, L.M.; Guthrie, B.L.; Nilsson, P.; Bosire, R.; Kiarie, J.; Farquhar, C.; Broliden, K.; Burgener, A.D. Genital injury signatures and microbiome alterations associated with depot medroxyprogesterone acetate usage and intravaginal drying practices. J. Infect. Dis. 2017, 215, 590–598. [Google Scholar] [CrossRef] [Green Version]
- Borgdorff, H.; Gautam, R.; Armstrong, S.D.; Xia, D.; Ndayisaba, G.F.; Van Teijlingen, N.H.; Geijtenbeek, T.B.H.; Wastling, J.M.; Van De Wijgert, J.H.H.M. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol. 2016, 9, 621–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalenskaya, I.A.; Chandra, N.; Yousefieh, N.; Fang, X.; Adedipe, O.E.; Jackson, S.S.; Anderson, S.M.; Mauck, C.K.; Schwartz, J.L.; Thurman, A.R.; et al. Use of contraceptive depot medroxyprogesterone acetate is associated with impaired cervicovaginal mucosal integrity. J. Clin. Investig. 2018, 128, 4622–4638. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 20 November 2020).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 20 November 2020).
- Anahtar, M.N.; Bowman, B.A.; Kwon, D.S. Efficient nucleic acid extraction and 16s rRNA gene sequencing for bacterial community characterization. J. Vis. Exp. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam, S.A.; McGarrell, D.M.; Garrity, G.M.; Tiedje, J.M. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005, 33, 294–296. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Anahtar, M.N.; Byrne, E.H.; Doherty, K.E.; Bowman, B.A.; Yamamoto, H.S.; Soumillon, M.; Padavattan, N.; Ismail, N.; Moodley, A.; Sabatini, M.E.; et al. Cervicovaginal Bacteria Are a Major Modulator of Host Inflammatory Responses in the Female Genital Tract. Immunity 2015, 42, 965–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Mcmurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edfeldt, G.; Lajoie, J.; Röhl, M.; Oyugi, J.; Åhlberg, A.; Khalilzadeh-Binicy, B.; Bradley, F.; Mack, M.; Kimani, J.; Omollo, K.; et al. Regular Use of Depot Medroxyprogesterone Acetate Causes Thinning of the Superficial Lining and Apical Distribution of Human Immunodeficiency Virus Target Cells in the Human Ectocervix. J. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamentsky, L.; Jones, T.R.; Fraser, A.; Bray, M.A.; Logan, D.J.; Madden, K.L.; Ljosa, V.; Rueden, C.; Eliceiri, K.W.; Carpenter, A.E. Improved structure, function and compatibility for cellprofiler: Modular high-throughput image analysis software. Bioinformatics 2011, 27, 1179–1180. [Google Scholar] [CrossRef] [Green Version]
- Saylor, J.; Ma, Z.; Goodridge, H.S.; Huang, F.; Cress, A.E.; Pandol, S.J.; Shiao, S.L.; Vidal, A.C.; Wu, L.; Nickols, N.G.; et al. Spatial Mapping of Myeloid Cells and Macrophages by Multiplexed Tissue Staining. Front. Immunol. 2018, 9, 2925. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Stassen, M.; Fondel, S.; Bopp, T.; Richter, C.; Müller, C.; Kubach, J.; Becker, C.; Knop, J.; Enk, A.H.; Schmitt, S.; et al. Human CD25+ regulatory T cells: Two subsets defined by the integrins α4β7 or α 4β1 confer distinct suppressive properties upon CD4+ T helper cells. Eur. J. Immunol. 2004, 34, 1303–1311. [Google Scholar] [CrossRef]
- Naranbhai, V.; Abdool Karim, S.S.; Altfeld, M.; Samsunder, N.; Durgiah, R.; Sibeko, S.; Abdool Karim, Q.; Carr, W.H. Innate immune activation enhances HIV acquisition in women, diminishing the effectiveness of tenofovir microbicide gel. J. Infect. Dis. 2012, 206, 993–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaul, R.; Prodger, J.; Joag, V.; Shannon, B.; Yegorov, S.; Galiwango, R.; McKinnon, L. Inflammation and HIV Transmission in Sub-Saharan Africa. Curr. HIV/AIDS Rep. 2015, 12, 216–222. [Google Scholar] [CrossRef]
- Masson, L.; Passmore, J.A.S.; Liebenberg, L.J.; Werner, L.; Baxter, C.; Arnold, K.B.; Williamson, C.; Little, F.; Mansoor, L.E.; Naranbhai, V.; et al. Genital Inflammation and the Risk of HIV Acquisition in Women. Clin. Infect. Dis. 2015, 61, 260–269. [Google Scholar] [CrossRef]
- Thibodeau, V.; Fourcade, L.; Labbé, A.C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Highly-Exposed HIV-1 seronegative Female Commercial Sex Workers sustain in their genital mucosa increased frequencies of tolerogenic myeloid and regulatory T-cells. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Barnitz, R.A.; Kreslavsky, T.; Brown, F.D.; Moffett, H.; Lemieux, M.E.; Kaygusuz, Y.; Meissner, T.; Holderried, T.A.W.; Chan, S.; et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 2015, 350, 334–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Ni, X.; Pan, X.; Lu, H.; Lu, Y.; Zhao, J.; Zheng, S.G.; Hippen, K.L.; Wang, X.; Lu, L. Human CD39hi regulatory T cells present stronger stability and function under inflammatory conditions. Cell. Mol. Immunol. 2017, 14, 521–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathaliyawala, T.; Kubota, M.; Yudanin, N.; Turner, D.; Camp, P.; Thome, J.J.C.; Bickham, K.L.; Lerner, H.; Goldstein, M.; Sykes, M.; et al. Distribution and Compartmentalization of Human Circulating and Tissue-Resident Memory T Cell Subsets. Immunity 2013, 38, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Shacklett, B.L.; Ferre, A.L.; Kiniry, B.E. Defining T Cell Tissue Residency in Humans: Implications for HIV Pathogenesis and Vaccine Design. Curr. HIV/AIDS Rep. 2020, 17, 109–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zevin, A.S.; Xie, I.Y.; Birse, K.; Arnold, K.; Romas, L.; Westmacott, G.; Novak, R.M.; Mccorrister, S.; Mckinnon, L.R. Microbiome Composition and Function Drives Wound-Healing Impairment in the Female Genital Tract. PLoS Pathog. 2016, 12, e1005889. [Google Scholar] [CrossRef] [Green Version]
- Schellenberg, J.J.; Links, M.G.; Hill, J.E.; Dumonceaux, T.J.; Kimani, J.; Jaoko, W.; Wachihi, C.; Mungai, J.N.; Peters, G.A.; Tyler, S.; et al. Molecular definition of vaginal microbiota in east african commercial sex workers. Appl. Environ. Microbiol. 2011, 77, 4066–4074. [Google Scholar] [CrossRef] [Green Version]
- De Jong, M.A.W.P.; De Witte, L.; Oudhoff, M.J.; Gringhuis, S.I.; Gallay, P.; Geijtenbeek, T.B.H. TNF-α and TLR agonists increase susceptibility to HIV-1 transmission by human Langerhans cells ex vivo. J. Clin. Investig. 2008, 118, 3440–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birse, K.; Arnold, K.B.; Novak, R.M.; Mccorrister, S.; Shaw, S.; Westmacott, G.R.; Ball, T.B.; Lauffenburger, D.A. Molecular Signatures of Immune Activation and Epithelial Barrier Remodeling Are Enhanced during the Luteal Phase of the Menstrual Cycle: Implications for HIV Susceptibility. J. Virol. 2015, 89, 8793–8805. [Google Scholar] [CrossRef] [Green Version]
- Woods, M.W.; Zahoor, M.A.; Dizzell, S.; Verschoor, C.P.; Kaushic, C. Medroxyprogesterone acetate-treated human, primary endometrial epithelial cells reveal unique gene expression signature linked to innate immunity and HIV-1 susceptibility. Am. J. Reprod. Immunol. 2018, 79, 12–14. [Google Scholar] [CrossRef]
- Dupont, H.A.; Lam, J.; Woods, M.W.; Zahoor, M.A.; Kaushic, C. Hormonal influence on HIV-1 transmission in the female genital tract: New insights from systems biology. Am. J. Reprod. Immunol. 2018, 80, 1–10. [Google Scholar] [CrossRef]
- Houshdaran, S.; Chen, J.C.; Vallvé-Juanico, J.; Balayan, S.; Vo, K.C.; Smith-McCune, K.; Greenblatt, R.M.; Irwin, J.C.; Giudice, L.C. Progestins related to progesterone and testosterone elicit divergent human endometrial transcriptomes and biofunctions. Int. J. Mol. Sci. 2020, 21, 2625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbuya, W.; McHaro, R.; Mhizde, J.; Mnkai, J.; Mahenge, A.; Mwakatima, M.; Mwalongo, W.; Chiwerengo, N.; Hölscher, M.; Lennemann, T.; et al. Depletion and activation of mucosal CD4 T cells in HIV infected women with HPVassociated lesions of the cervix uteri. PLoS ONE 2020, 15, e0240154. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.; Fraser, I.S.; Baker, J.; Archer, D.; Landgren, B.M.; Killick, S.; Soutter, P.; Krause, T.; D’Arcangues, C. A randomized comparison of the effects on vaginal and cervical epithelium of a placebo vaginal ring with non-use of a ring. Contraception 2000, 62, 83–89. [Google Scholar] [CrossRef]
- Posavad, C.M.; Zhao, L.; Dong, L.; Jin, L.; Stevens, C.E.; Magaret, A.S.; Johnston, C.; Wald, A.; Zhu, J.; Corey, L.; et al. Enrichment of herpes simplex virus type 2 (HSV-2) reactive mucosal T cells in the human female genital tract. Mucosal Immunol. 2017, 10, 1259–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, E.; Yifei, M.; Farhat, S.; Shiboski, F.; Moscicki, A. Covariates of Cervical Cytokine mRNA Expression by Real-Time PCR in Adolescents and Young Women: Effects of Chlamydia trachomatis Infection, Hormonal Contraception, and Smoking. J. Clin. Immunol. Immunol. 2006, 99, 222–229. [Google Scholar] [CrossRef]
- McKinnon, L.R.; Izulla, P.; Nagelkerke, N.; Munyao, J.; Wanjiru, T.; Shaw, S.Y.; Gichuki, R.; Kariuki, C.; Muriuki, F.; Musyoki, H.; et al. Risk Factors for HIV Acquisition in a Prospective Nairobi-Based Female Sex Worker Cohort. AIDS Behav. 2015, 19, 2204–2213. [Google Scholar] [CrossRef]







| Parameter | HESN (n = 29) | NN (n = 33) | p-Value |
|---|---|---|---|
| Median or Number (Range or %) | Median or Number (Range or %) | ||
| Age (years) | 37 (30–50) | 31 (21–47) | 0.001 |
| Years since last pregnancy a | 7 (1–19) | 8 (5–22) | ns |
| Progesterone levels (ng/mL) b | 2.6 (0.1–17) | 3.9 (0.1–16.4) | ns |
| BV c | |||
| Negative (0–3) | 14 (48%) | 17 (53%) | ns |
| Intermediate (4–6) | 9 (31%) | 6 (19%) | ns |
| Positive (7–10) | 6 (21%) | 9 (28%) | ns |
| Yeast infection | 1 (3%) | 1 (3%) | ns |
| No. of clients last 7 days | 5 (0–50) | 4.5 (0–30) | ns |
| Unprotected sex acts last 7 days d | 0 (0–6) | 0 (0–1) | ns |
| Vaginal douching e | 13 (45%) | 8 (24%) | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röhl, M.; Tjernlund, A.; Lajoie, J.; Edfeldt, G.; Bradley, F.; Bergström, S.; Kaldhusdal, V.; Åhlberg, A.; Månberg, A.; Omollo, K.; et al. HIV-Exposed Seronegative Sex Workers Express Low T-Cell Activation and an Intact Ectocervical Tissue Microenvironment. Vaccines 2021, 9, 217. https://doi.org/10.3390/vaccines9030217
Röhl M, Tjernlund A, Lajoie J, Edfeldt G, Bradley F, Bergström S, Kaldhusdal V, Åhlberg A, Månberg A, Omollo K, et al. HIV-Exposed Seronegative Sex Workers Express Low T-Cell Activation and an Intact Ectocervical Tissue Microenvironment. Vaccines. 2021; 9(3):217. https://doi.org/10.3390/vaccines9030217
Chicago/Turabian StyleRöhl, Maria, Annelie Tjernlund, Julie Lajoie, Gabriella Edfeldt, Frideborg Bradley, Sofia Bergström, Vilde Kaldhusdal, Alexandra Åhlberg, Anna Månberg, Kenneth Omollo, and et al. 2021. "HIV-Exposed Seronegative Sex Workers Express Low T-Cell Activation and an Intact Ectocervical Tissue Microenvironment" Vaccines 9, no. 3: 217. https://doi.org/10.3390/vaccines9030217
APA StyleRöhl, M., Tjernlund, A., Lajoie, J., Edfeldt, G., Bradley, F., Bergström, S., Kaldhusdal, V., Åhlberg, A., Månberg, A., Omollo, K., Boily-Larouche, G., Asghar, M., Kwon, D. S., Oyugi, J., Kimani, J., Nilsson, P., Fowke, K. R., & Broliden, K. (2021). HIV-Exposed Seronegative Sex Workers Express Low T-Cell Activation and an Intact Ectocervical Tissue Microenvironment. Vaccines, 9(3), 217. https://doi.org/10.3390/vaccines9030217