Protective Effect of Nasal Colonisation with ∆cps/piaA and ∆cps/proABCStreptococcus pneumoniae Strains against Recolonisation and Invasive Infection
Abstract
:1. Introduction
2. Methods
2.1. Bacterial Methods and Construction of the Deletion Mutant Strains
2.2. C3b Complement Binding Assays
2.3. Immunofluorescence Microscopy
2.4. Immunological Assays
2.5. Protein Microarray Assays
2.6. Mouse Experimental Models
2.7. RNA Samples and Sequencing
2.8. Statistical Analyses
3. Results
3.1. Design and Characterisation of Unencapsulated Mutant Strains
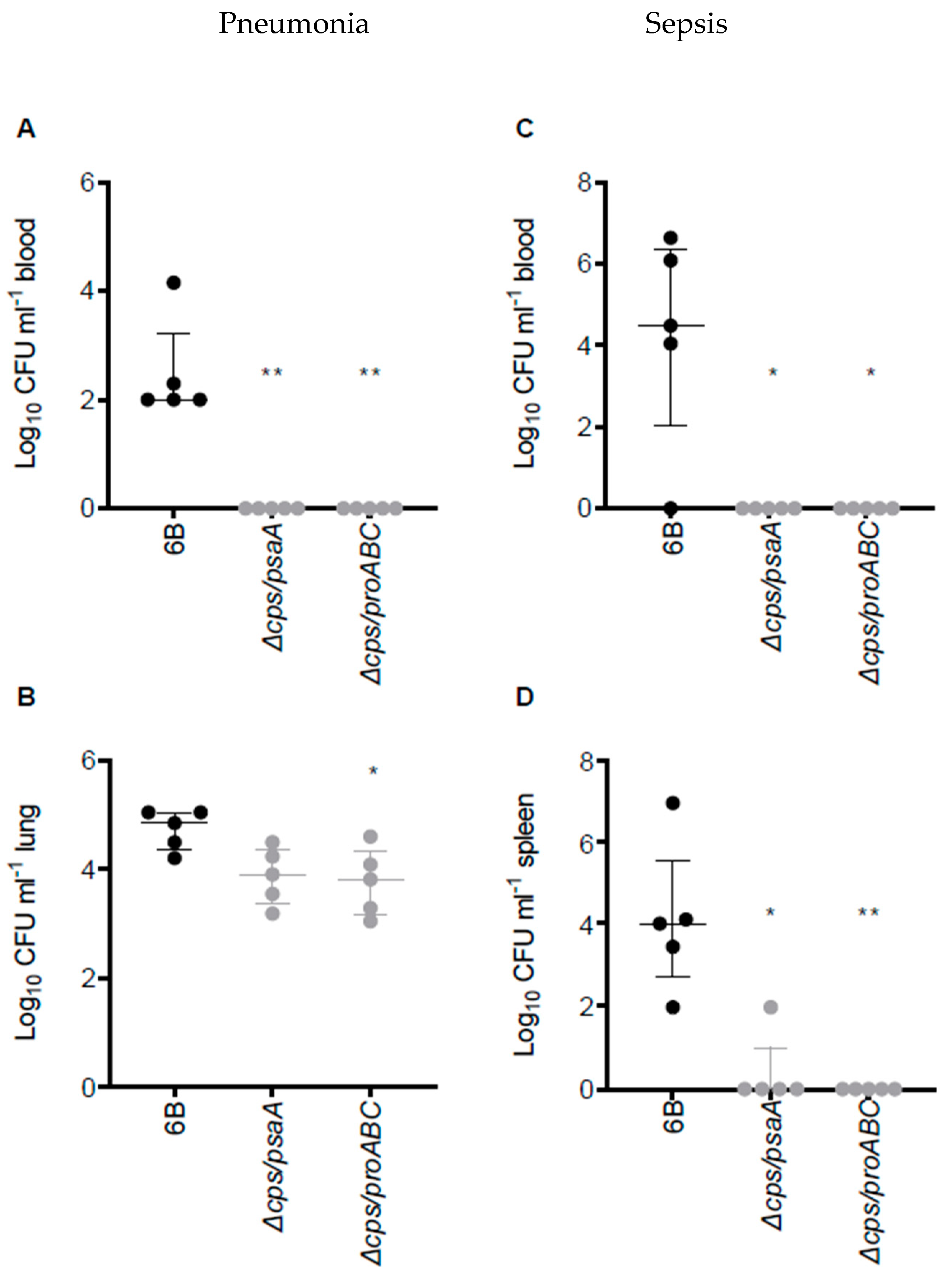
3.2. Virulence of ∆cps/psaA and ∆cps/proABC Strains
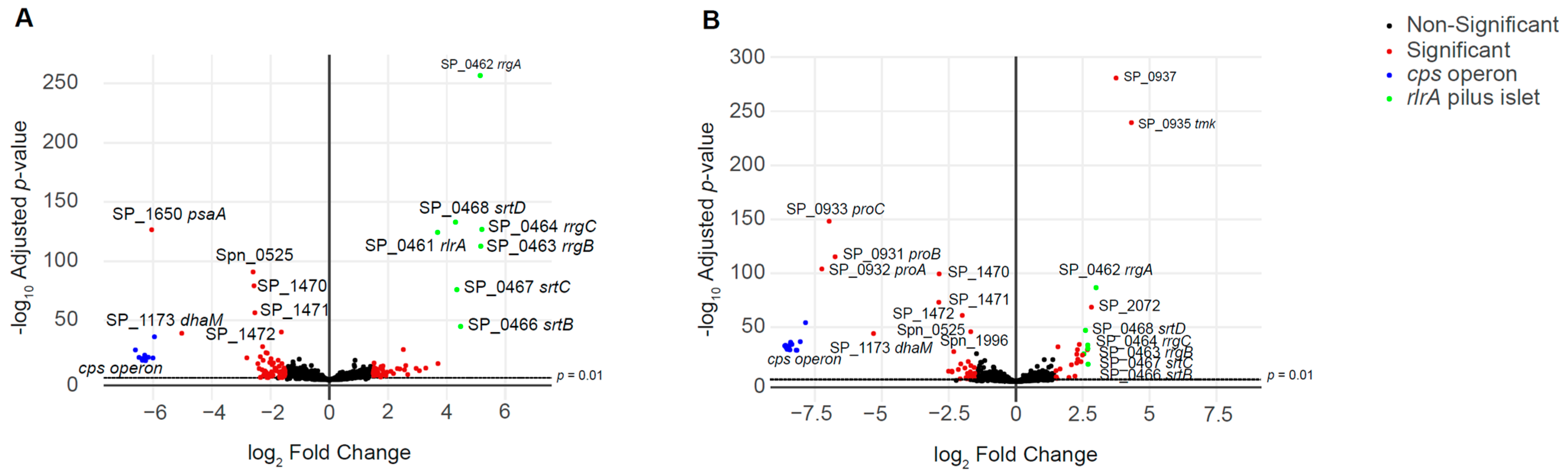
3.3. RNA-seq Analysis of the Unencapsulated Double Mutant Strains
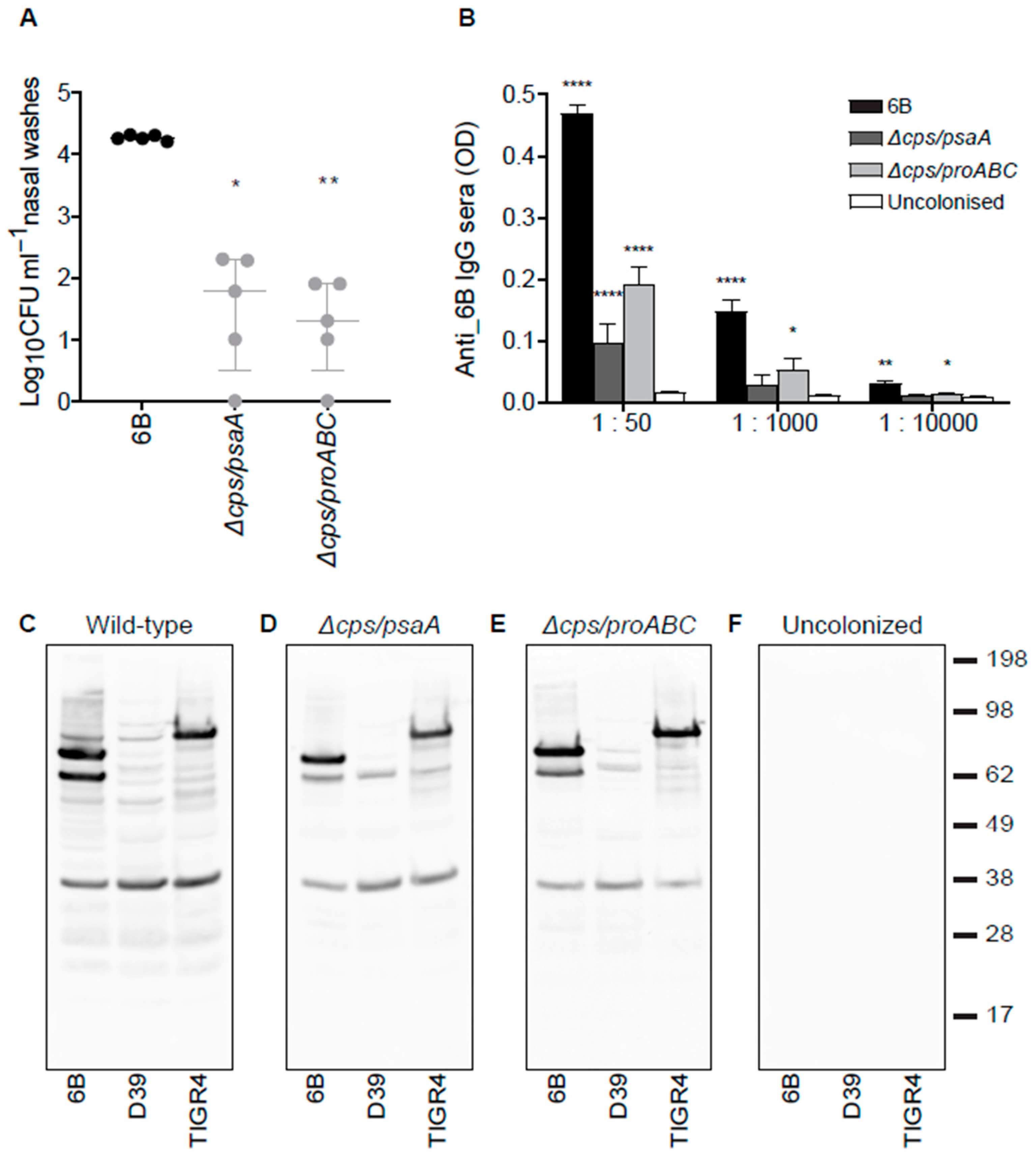
3.4. Nasopharyngeal Colonisation by Double Mutant Strains and Subsequent Serological Responses
3.5. Identification of Protein Antigens Recognised by Serological Responses to Colonisation Using Protein Microarrays
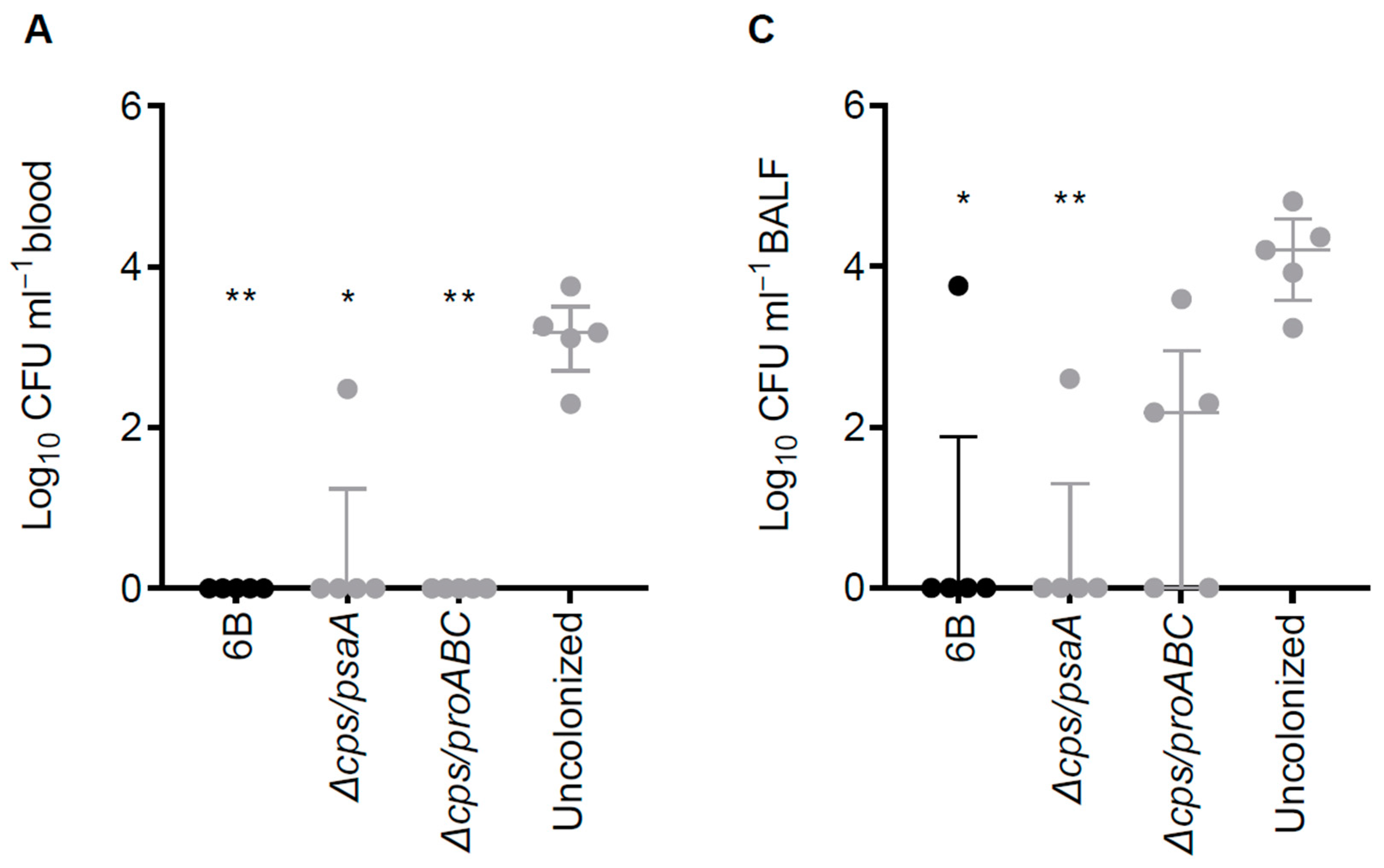
3.6. Double Mutant Colonisation Protects against Bacteraemia during Pneumonia Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, W.S.; Macfarlane, J.T.; Boswell, T.C.; Harrison, T.G.; Rose, D.; Leinonen, M.; Saikku, P. Study of community acquired pneumonia aetiology (SCAPA) in adults admitted to hospital: Implications for management guidelines. Thorax 2001, 56, 296–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melegaro, A.; Edmunds, W.J.; Pebody, R.; Miller, E.; George, R. The current burden of pneumococcal disease in England and Wales. J. Infect. 2006, 52, 37–48. [Google Scholar] [CrossRef]
- Garcha, D.S.; Thurston, S.J.; Patel, A.R.C.; Mackay, A.J.; Goldring, J.J.P.; Donaldson, G.C.; McHugh, T.D.; Wedzicha, J.A. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 2012, 67, 1075–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moberley, S.; Holden, J.; Tatham, D.P.; Andrews, R.M. Vaccines for preventing pneumococcal infection in adults. Cochrane Database Syst. Rev. 2013, 2013, CD000422. [Google Scholar] [CrossRef]
- Bonten, M.J.; Huijts, S.M.; Bolkenbaas, M.; Webber, C.; Patterson, S.; Gault, S.; Van Werkhoven, C.H.; Van Deursen, A.M.; Sanders, E.A.; Verheij, T.J.; et al. Polysaccharide conjugate vaccine against pneumococcal pneumonia in adults. N. Engl. J. Med. 2015, 372, 1114–1125. [Google Scholar] [CrossRef] [Green Version]
- Abdullahi, O.; Karani, A.; Tigoi, C.C.; Mugo, D.; Kungu, S.; Wanjiru, E.; Jomo, J.; Musyimi, R.; Lipsitch, M.; Scott, J.A.G. The prevalence and risk factors for pneumococcal colonization of the nasopharynx among children in Kilifi District, Kenya. PLoS ONE 2012, 7, e30787. [Google Scholar] [CrossRef]
- Miller, E.; Andrews, N.J.; Waight, P.A.; Slack, M.P.; George, R.C. Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: An observational cohort study. Lancet Infect. Dis. 2011, 11, 760–768. [Google Scholar] [CrossRef]
- Balsells, E.; Guillot, L.; Nair, H.; Kyaw, M.H. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0177113. [Google Scholar] [CrossRef]
- Bradshaw, J.L.; Pipkins, H.R.; Keller, L.E.; Pendarvis, J.K.; McDaniel, L.S. Mucosal infections and invasive potential of nonencapsulated Streptococcus pneumoniae are enhanced by oligopeptide binding proteins AliC and AliD. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lochen, A.; Croucher, N.J.; Anderson, R.M. Divergent serotype replacement trends and increasing diversity in pneumococcal disease in high income settings reduce the benefit of expanding vaccine valency. Sci. Rep. 2020, 10, 18977. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Cohen, J.M.; Reglinski, M.; José, R.J.; Chan, W.Y.; Marshall, H.; De Vogel, C.; Gordon, S.; Goldblatt, D.; Petersen, F.C.; et al. Naturally acquired human immunity to pneumococcus is dependent on antibody to protein antigens. PLoS Pathog. 2017, 13, e1006137. [Google Scholar] [CrossRef]
- Lipsitch, M.; Whitney, C.G.; Zell, E.; Kaijalainen, T.; Dagan, R.; Malley, R. Are anticapsular antibodies the primary mechanism of protection against invasive pneumococcal disease? PLoS Med. 2005, 2, e15. [Google Scholar] [CrossRef]
- Cohen, J.M.; Wilson, R.; Shah, P.; Baxendale, H.E.; Brown, J.S. Lack of cross-protection against invasive pneumonia caused by heterologous strains following murine Streptococcus pneumoniae nasopharyngeal colonisation despite whole cell ELISAs showing significant cross-reactive IgG. Vaccine 2013, 31, 2328–2332. [Google Scholar] [CrossRef]
- Cohen, J.M.; Chimalapati, S.; de Vogel, C.; van Belkum, A.; Baxendale, H.E.; Brown, J.S. Contributions of capsule, lipoproteins and duration of colonisation towards the protective immunity of prior Streptococcus pneumoniae nasopharyngeal colonisation. Vaccine 2012, 30, 4453–4459. [Google Scholar] [CrossRef] [Green Version]
- Roche, A.M.; King, S.J.; Weiser, J.N. Live attenuated Streptococcus pneumoniae strains induce serotype-independent mucosal and systemic protection in mice. Infect. Immun. 2007, 75, 2469–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, A.K.A.; Ferreira, D.M.; Gritzfeld, J.F.; Wright, A.D.; Armitage, K.; Jambo, K.C.; Bate, E.; El Batrawy, S.; Collins, A.; Gordon, S.B. Human nasal challenge with Streptococcus pneumoniae is immunising in the absence of carriage. PLoS Pathog. 2012, 8, e1002622. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.K.A.; Bangert, M.; Gritzfeld, J.F.; Ferreira, D.M.; Jambo, K.C.; Wright, A.D.; Collins, A.M.; Gordon, S.B. Experimental human pneumococcal carriage augments IL-17A-dependent T-cell defence of the lung. PLoS Pathog. 2013, 9, e1003274. [Google Scholar] [CrossRef] [PubMed]
- David, S.C.; Laan, Z.; Minhas, V.; Chen, A.Y.; Davies, J.; Hirst, T.R.; McColl, S.R.; Alsharifi, M.; Paton, J.C. Enhanced safety and immunogenicity of a pneumococcal surface antigen A mutant whole-cell inactivated pneumococcal vaccine. Immunol. Cell Biol. 2019, 97, 726–739. [Google Scholar] [CrossRef]
- McCool, T.L.; Cate, T.R.; Moy, G.; Weiser, J.N. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 2002, 195, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.; Cohen, J.M.; Jose, R.J.; de Vogel, C.; Baxendale, H.; Brown, J.S. Protection against Streptococcus pneumoniae lung infection after nasopharyngeal colonization requires both humoral and cellular immune responses. Mucosal Immunol. 2015, 8, 627–639. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Clarke, T.B.; Weiser, J.N. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J. Clin. Investig. 2009, 119, 1899–1909. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.M.; Khandavilli, S.; Camberlein, E.; Hyams, C.; Baxendale, H.E.; Brown, J.S. Protective contributions against invasive Streptococcus pneumoniae pneumonia of antibody and Th17-cell responses to nasopharyngeal colonisation. PLoS ONE 2011, 6, e25558. [Google Scholar] [CrossRef]
- Ramos-Sevillano, E.; Ercoli, G.; Felgner, P.; De Assis, R.R.; Nakajima, R.; Goldblatt, D.; Heyderman, R.S.; Gordon, S.B.; Ferreira, D.M.; Brown, J.S. Preclinical development of virulence attenuated Streptococcus pneumoniae strains able to enhance protective immunity against pneumococcal infection. Am. J. Respir. Crit. Care Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.M.; Neill, D.R.; Bangert, M.; Gritzfeld, J.F.; Green, N.; Wright, A.K.A.; Pennington, S.H.; Moreno, L.B.; Moreno, A.T.; Miyaji, E.N.; et al. Controlled human infection and rechallenge with Streptococcus pneumoniae reveals the protective efficacy of carriage in healthy adults. Am. J. Respir. Crit. Care Med. 2013, 187, 855–864. [Google Scholar] [CrossRef] [Green Version]
- Jochems, S.P.; De Ruiter, K.; Solórzano, C.; Voskamp, A.; Mitsi, E.; Nikolaou, E.; Carniel, B.F.; Pojar, S.; German, E.L.; Reiné, J.; et al. Innate and adaptive nasal mucosal immune responses following experimental human pneumococcal colonization. J. Clin. Investig. 2019, 129, 4523–4538. [Google Scholar] [CrossRef] [Green Version]
- Chimalapati, S.; Cohen, J.; Camberlein, E.; Durmort, C.; Baxendale, H.; De Vogel, C.; Van Belkum, A.; Brown, J.S. Infection with conditionally virulent Streptococcus pneumoniae ∆pab strains induces antibody to conserved protein antigens but does not protect against systemic infection with heterologous strains. Infect. Immun. 2011, 79, 4965–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Seon, S.H.; Luong, T.T.; Ghosh, P.; Pyo, S.; Rhee, D.K. Immunization with attenuated non-transformable pneumococcal pep27 and comD mutant provides serotype-independent protection against pneumococcal infection. Vaccine 2019, 37, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Rosch, J.W.; Iverson, A.R.; Humann, J.; Mann, B.; Gao, G.; Vogel, P.; Mina, M.; Murrah, K.A.; Perez, A.C.; Swords, W.E.; et al. A live-attenuated pneumococcal vaccine elicits CD4+ T-cell dependent class switching and provides serotype independent protection against acute otitis media. EMBO Mol. Med. 2014, 6, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Chimalapati, S.; Cohen, J.M.; Camberlein, E.; Macdonald, N.; Durmort, C.; Vernet, T.; Hermans, P.W.M.; Mitchell, T.; Brown, J.S. Effects of deletion of the Streptococcus pneumoniae lipoprotein diacylglyceryl transferase gene lgt on ABC transporter function and on growth in vivo. PLoS ONE 2012, 7, e41393. [Google Scholar] [CrossRef] [Green Version]
- Berry, A.M.; Paton, J.C. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 1996, 64, 5255–5262. [Google Scholar] [CrossRef] [Green Version]
- Tseng, H.J.; McEwan, A.G.; Paton, J.C.; Jennings, M.P. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect. Immun. 2002, 70, 1635–1639. [Google Scholar] [CrossRef] [Green Version]
- van Opijnen, T.; Camilli, A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012, 22, 2541–2551. [Google Scholar] [CrossRef] [Green Version]
- Havarstein, L.S.; Coomaraswamy, G.; Morrison, D.A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 1995, 92, 11140–11144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyams, C.; Camberlein, E.; Cohen, J.M.; Bax, K.; Brown, J.S. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 2010, 78, 704–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reglinski, M.; Ercoli, G.; Plumptre, C.; Kay, E.; Petersen, F.C.; Paton, J.C.; Wren, B.W.; Brown, J.S. A recombinant conjugated pneumococcal vaccine that protects against murine infections with a similar efficacy to Prevnar-13. NPJ Vaccines 2018, 3, 53. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.-Y.; Entwisle, C.; Ercoli, G.; Ramos-Sevillano, E.; McIlgorm, A.; Cecchini, P.; Bailey, C.; Lam, O.; Whiting, G.; Green, N.; et al. A Novel, multiple-antigen pneumococcal vaccine protects against lethal Streptococcus pneumoniae challenge. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [Green Version]
- Croucher, N.J.; Campo, J.J.; Le, T.Q.; Liang, X.; Bentley, S.D.; Hanage, W.P.; Lipsitch, M. Diverse evolutionary patterns of pneumococcal antigens identified by pangenome-wide immunological screening. Proc. Natl. Acad. Sci. USA 2017, 114, E357–E366. [Google Scholar] [CrossRef] [Green Version]
- Ercoli, G.; Ramos-Sevillano, E.; Nakajima, R.; de Assis, R.R.; Jasinskas, A.; Goldblatt, D.; Felgner, P.; Weckbecker, G.; Brown, J. The influence of B cell depletion therapy on naturally acquired immunity to Streptococcus pneumoniae. Front. Immunol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Brooks, L.R.K.; Mias, G.I. Streptococcus pneumoniae’s virulence and host immunity: Aging, diagnostics, and prevention. Front. Immunol. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Bidossi, A.; Mulas, L.; Decorosi, F.; Colomba, L.; Ricci, S.; Pozzi, G.; Deutscher, J.; Viti, C.; Oggioni, M.R. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 2012, 7, e33320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campo, J.J.; Le, T.Q.; Pablo, J.V.; Hung, C.; Teng, A.A.; Tettelin, H.; Tate, A.; Hanage, W.P.; Alderson, M.R.; Liang, X.; et al. Panproteome-wide analysis of antibody responses to whole cell pneumococcal vaccination. Elife 2018, 7, 7. [Google Scholar] [CrossRef]
- Goldblatt, D.; Hussain, M.; Andrews, N.; Ashton, L.; Virta, C.; Melegaro, A.; Pebody, R.; George, R.; Soininen, A.; Edmunds, J.; et al. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: A longitudinal household study. J. Infect. Dis. 2005, 192, 387–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granat, S.M.; Ollgren, J.; Herva, E.; Mia, Z.; Auranen, K.; Makela, P.H. Epidemiological evidence for serotype-independent acquired immunity to pneumococcal carriage. J. Infect. Dis. 2009, 200, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberger, D.M.; Dagan, R.; Givon-Lavi, N.; Regev-Yochay, G.; Malley, R.; Lipsitch, M. Epidemiologic evidence for serotype-specific acquired immunity to pneumococcal carriage. J. Infect. Dis. 2008, 197, 1511–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobey, S.; Lipsitch, M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science 2012, 335, 1376–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.S. Improving Pulmonary Immunity to Bacterial Pathogens through Streptococcus pneumoniae Colonization of the Nasopharynx. Am. J. Respir. Crit. Care Med. 2020, 201, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Mitsi, E.; Carniel, B.; Reiné, J.; Rylance, J.; Zaidi, S.; Soares-Schanoski, A.; Connor, V.; Collins, A.M.; Schlitzer, A.; Nikolaou, E.; et al. Nasal pneumococcal density is associated with microaspiration and heightened human alveolar macrophage responsiveness to bacterial pathogens. Am. J. Respir. Crit. Care Med. 2020, 201, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Gilliland, S.M.; Holden, D.W. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 2001, 40, 572–585. [Google Scholar] [CrossRef]
- Belitsky, B.R.; Brill, J.; Bremer, E.; Sonenshein, A.L. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 2001, 183, 4389–4392. [Google Scholar] [CrossRef] [Green Version]
- Basset, A.; Trzcinski, K.; Hermos, C.; O’Brien, K.L.; Reid, R.; Santosham, M.; McAdam, A.J.; Lipsitch, M.; Malley, R. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 2007, 45, 1684–1689. [Google Scholar] [CrossRef] [Green Version]
- LeMieux, J.; Hava, D.L.; Basset, A.; Camilli, A. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 2006, 74, 2453–2456. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| BHN418 Gene Number and Name or Operon Function | TIGR4 Gene Number (If Known) | ∆cps/psaA | ∆cps/proABC |
|---|---|---|---|
| Upregulated genes | |||
| Spn_00124_xylB | SP_1855 | 2.956 | 1.538 |
| Spn_00125_6 regulator/cation efflux | SP_1856-7 | 3.493 (0.293) | 2.099 (0.159 |
| Spn_00599-00 Blp bacteriocin | SP_0041- | 1.838 (0.127) | |
| Spn_00677_mutT | SP_0119 | 1.533 | 1.502 |
| Spn_00914_aliA | SP_0366 | 2.520 | 2.677 |
| Spn_00963-68 fatty acid synthesis | SP_0415-419-420 | 1.623 (0.153) | |
| Spn_01010-15_rlrA islet | SP_0462-68 | 4.766 (0.438) | 2.706 (0.157) |
| Spn_01098-00 Blp bacteriocin | SP_0531-33 | 2.168 (0.510) | |
| Spn_01108_blpX | SP_0544 | 2.561 | |
| Spn_01111 Blp bacteriocin | SP_0547 | 1.506 | |
| Spn_02091-97 fructose PTS | SP_1615-21 | 2.354 (0.083) | |
| Downregulated genes | |||
| Spn_00121-22 Galactose metabolism | SP_1852-53 | −2.058 (0.134) | −1.690 (0.006) |
| Spn_00617_bgaC | SP_0060 | −1.698 | |
| Spn_00618-21 sugar PTS | SP_0060-64 | −2.158 (0.238) | −2.273 (0.317) |
| Spn_00641-42 glycerol ABC transporter | SP_0091-92 | −2.344 (0.125) | −1.669 (0.183) |
| Spn_00899-913 cps locus | SP_0343-65 | −6.304 (0.164) | −8.385 (0.220) |
| Spn_01042-46 Endo-beta-N-acetylglucosaminidase | SP_0498 | −1.771 (0.331) | |
| Spn_01479-81_proBAC | SP_0931-33 | −6.992 (0.247) | |
| Spn_01723-26_lacGEFT | SP_1184-87 | −1.978 (0.281) | −1.831 (0.023) |
| Spn_02120_psaA | SP_1650 | −6.048 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Sevillano, E.; Ercoli, G.; Guerra-Assunção, J.A.; Felgner, P.; Ramiro de Assis, R.; Nakajima, R.; Goldblatt, D.; Tetteh, K.K.A.; Heyderman, R.S.; Gordon, S.B.; et al. Protective Effect of Nasal Colonisation with ∆cps/piaA and ∆cps/proABCStreptococcus pneumoniae Strains against Recolonisation and Invasive Infection. Vaccines 2021, 9, 261. https://doi.org/10.3390/vaccines9030261
Ramos-Sevillano E, Ercoli G, Guerra-Assunção JA, Felgner P, Ramiro de Assis R, Nakajima R, Goldblatt D, Tetteh KKA, Heyderman RS, Gordon SB, et al. Protective Effect of Nasal Colonisation with ∆cps/piaA and ∆cps/proABCStreptococcus pneumoniae Strains against Recolonisation and Invasive Infection. Vaccines. 2021; 9(3):261. https://doi.org/10.3390/vaccines9030261
Chicago/Turabian StyleRamos-Sevillano, Elisa, Giuseppe Ercoli, José Afonso Guerra-Assunção, Philip Felgner, Rafael Ramiro de Assis, Rie Nakajima, David Goldblatt, Kevin Kweku Adjei Tetteh, Robert Simon Heyderman, Stephen Brian Gordon, and et al. 2021. "Protective Effect of Nasal Colonisation with ∆cps/piaA and ∆cps/proABCStreptococcus pneumoniae Strains against Recolonisation and Invasive Infection" Vaccines 9, no. 3: 261. https://doi.org/10.3390/vaccines9030261
APA StyleRamos-Sevillano, E., Ercoli, G., Guerra-Assunção, J. A., Felgner, P., Ramiro de Assis, R., Nakajima, R., Goldblatt, D., Tetteh, K. K. A., Heyderman, R. S., Gordon, S. B., Ferreria, D. M., & Brown, J. S. (2021). Protective Effect of Nasal Colonisation with ∆cps/piaA and ∆cps/proABCStreptococcus pneumoniae Strains against Recolonisation and Invasive Infection. Vaccines, 9(3), 261. https://doi.org/10.3390/vaccines9030261






