Construction of Porcine Epidemic Diarrhea Virus-Like Particles and Its Immunogenicity in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Plasmids
2.3. Production and Purification of VLPs
2.4. Western Blot Analysis
2.5. Mice Immunization
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Serum Neutralization (SN) Test
2.8. Statistical Analyses
3. Results
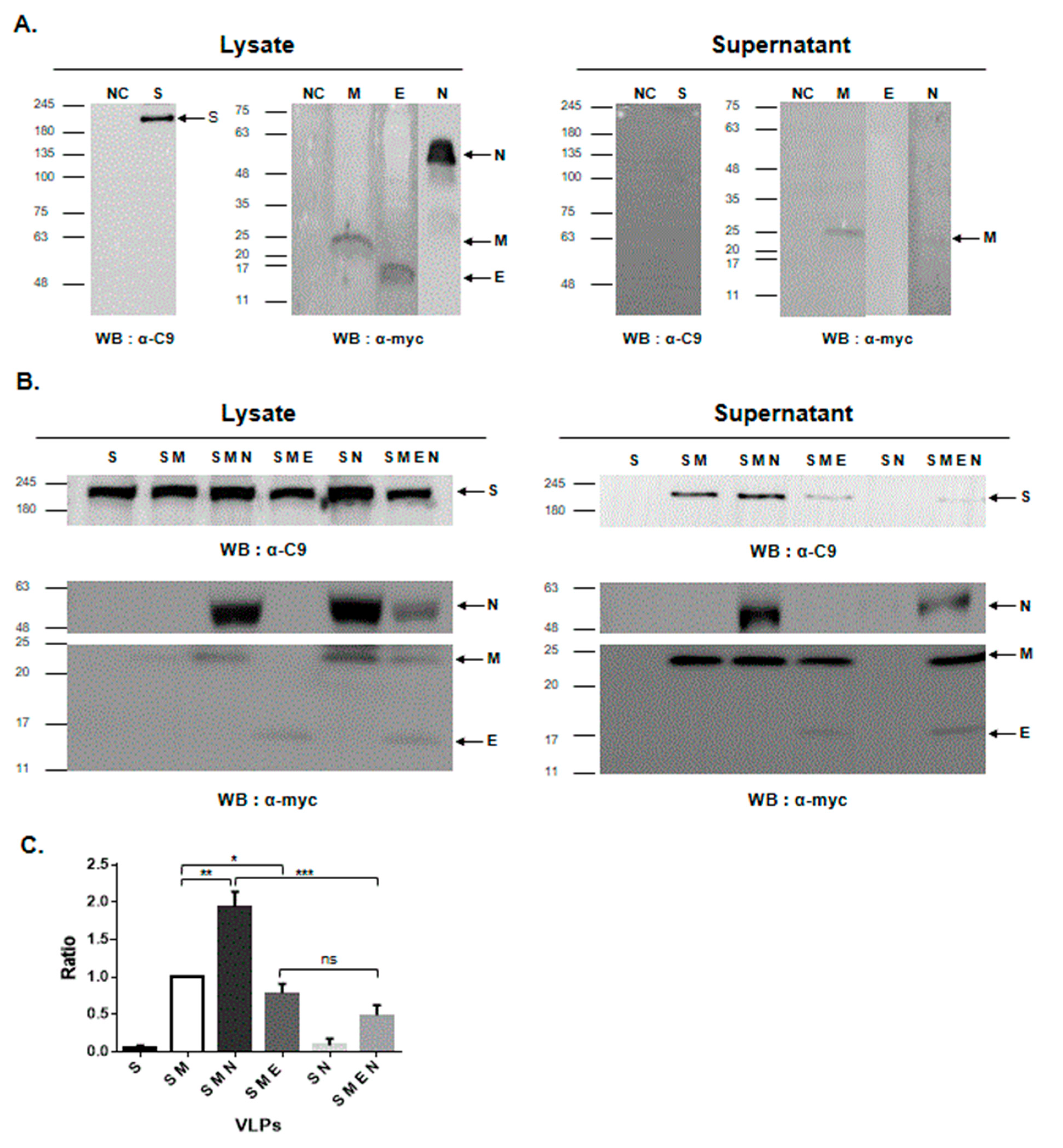
3.1. M Protein Expression Is Sufficient for the Formation of PED VLPs
3.2. PED VLPs Induce Strong Immune Responses in Mice
3.3. PED VLP Immunization Induced Neutralizing Antibodies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duarte, M.; Gelfi, J.; Lambert, P.; Rasschaert, D.; Laude, H. Genome organization of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1993, 342, 55–60. [Google Scholar]
- Debouck, P.; Pensaert, M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980, 41, 219–223. [Google Scholar]
- Turgeon, D.C.; Morin, M.; Jolette, J.; Higgins, R.; Marsolais, G.; DiFranco, E. Coronavirus-like particles associated with diarrhea in baby pigs in Quebec. Can. Vet. J. 1980, 21, 100-xxiii. [Google Scholar] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kweon, C.; Kwon, B.; Jung, T.; Kee, Y.; Hur, D.; Hwang, E.; Rhee, J.; An, S. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J. Vet. Res. 1993, 33, 249–254. [Google Scholar]
- Sueyoshi, M.; Tsuda, T.; Yamazaki, K.; Yoshida, K.; Nakazawa, M.; Sato, K.; Minami, T.; Iwashita, K.; Watanabe, M.; Suzuki, Y. An immunohistochemical investigation of porcine epidemic diarrhoea. J. Comp. Pathol. 1995, 113, 59–67. [Google Scholar] [CrossRef]
- Lee, H.M.; Lee, B.J.; Tae, J.H.; Kweon, C.H.; Lee, Y.S.; Park, J.H. Detection of porcine epidemic diarrhea virus by immunohistochemistry with recombinant antibody produced in phages. J. Vet. Med. Sci. 2000, 62, 333–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.W.; Dickerman, A.W.; Pineyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef]
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 2001, 23, 137–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Cruz, D.J.; Shin, H.J. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 2011, 156, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tang, J.; Ma, Y.; Liang, X.; Yang, Y.; Peng, G.; Qi, Q.; Jiang, S.; Li, J.; Du, L.; et al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015, 89, 6121–6125. [Google Scholar] [CrossRef] [Green Version]
- Kirchdoerfer, R.N.; Bhandari, M.; Martini, O.; Sewall, L.M.; Bangaru, S.; Yoon, K.J.; Ward, A.B. Structure and immune recognition of the porcine epidemic diarrhea virus spike protein. Structure 2020. [Google Scholar] [CrossRef]
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534. [Google Scholar] [CrossRef] [Green Version]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Venkatagopalan, P.; Daskalova, S.M.; Lopez, L.A.; Dolezal, K.A.; Hogue, B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology 2015, 478, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Torres, J.L.; Dediego, M.L.; Alvarez, E.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef] [Green Version]
- de Haan, C.A.; Rottier, P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005, 64, 165–230. [Google Scholar]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Arriba, M.L.; Carvajal, A.; Pozo, J.; Rubio, P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet. Immunol. Immunopathol. 2002, 85, 85–97. [Google Scholar] [CrossRef]
- Lee, S.H.; Yang, D.K.; Kim, H.H.; Cho, I.S. Efficacy of inactivated variant porcine epidemic diarrhea virus vaccines in growing pigs. Clin. Exp. Vaccine Res. 2018, 7, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Song, D.S.; Oh, J.S.; Kang, B.K.; Yang, J.S.; Moon, H.J.; Yoo, H.S.; Jang, Y.S.; Park, B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007, 82, 134–140. [Google Scholar] [CrossRef]
- Won, H.; Lim, J.; Noh, Y.H.; Yoon, I.; Yoo, H.S. Efficacy of Porcine Epidemic Diarrhea Vaccines: A Systematic Review and Meta-Analysis. Vaccines 2020, 8, 642. [Google Scholar] [CrossRef]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017, 206, 45–51. [Google Scholar] [CrossRef]
- Crawford, K.; Lager, K.M.; Kulshreshtha, V.; Miller, L.C.; Faaberg, K.S. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res. 2016, 226, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.; Singh, P.; Pillatzki, A.; Nelson, E.; Webb, B.; Dillberger-Lawson, S.; Ramamoorthy, S. A Minimally Replicative Vaccine Protects Vaccinated Piglets Against Challenge With the Porcine Epidemic Diarrhea Virus. Front. Vet. Sci. 2019, 6, 347. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef]
- Noad, R.; Roy, P. Virus-like particles as immunogens. Trends Microbiol. 2003, 11, 438–444. [Google Scholar] [CrossRef]
- Hofmann, M.; Wyler, R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988, 26, 2235–2239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Kang, K.J.; Ryu, J.H.; Park, J.Y.; Jang, H.; Sung, D.J.; Kang, J.G.; Shin, H.J. Porcine epidemic diarrhea vaccine evaluation using a newly isolated strain from Korea. Vet. Microbiol. 2018, 221, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Lin, Y.; Qin, J.; Li, Q.; Xue, C.; Cao, Y. Neutralizing antibodies against porcine epidemic diarrhea virus block virus attachment and internalization. Virol. J. 2018, 15, 1–9. [Google Scholar] [CrossRef]
- Vennema, H.; Godeke, G.J.; Rossen, J.W.; Voorhout, W.F.; Horzinek, M.C.; Opstelten, D.J.; Rottier, P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996, 15, 2020–2028. [Google Scholar] [CrossRef]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.; et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.K.; Chang, S.C.; Huang, C.C.; Lee, T.T.; Hsiao, C.W.; Kou, Y.H.; Chen, I.Y.; Chang, C.K.; Huang, T.H.; Chang, M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005, 79, 13848–13855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zheng, X.; Gai, W.; Zhao, Y.; Wang, H.; Wang, H.; Feng, N.; Chi, H.; Qiu, B.; Li, N.; et al. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget 2017, 8, 12686–12694. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Wang, C.; Yan, F.; Zheng, X.; Wang, H.; Jin, H.; Wang, C.; Zhao, Y.; Feng, N.; Wang, T.; Gao, Y.; et al. Porcine epidemic diarrhea virus virus-like particles produced in insect cells induce specific immune responses in mice. Virus Genes 2017, 53, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Syomin, B.V.; Ilyin, Y.V. Virus-Like Particles as an Instrument of Vaccine Production. Mol. Biol. 2019, 53, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Ruch, T.R.; Machamer, C.E. The coronavirus E protein: Assembly and beyond. Viruses 2012, 4, 363–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Cao, J.; Zhao, L.J.; Qin, Z.L.; Ke, J.S.; Pan, W.; Ren, H.; Yu, J.G.; Qi, Z.T. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology 2005, 331, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Yoon, J.; Park, J.-E. Construction of Porcine Epidemic Diarrhea Virus-Like Particles and Its Immunogenicity in Mice. Vaccines 2021, 9, 370. https://doi.org/10.3390/vaccines9040370
Kim J, Yoon J, Park J-E. Construction of Porcine Epidemic Diarrhea Virus-Like Particles and Its Immunogenicity in Mice. Vaccines. 2021; 9(4):370. https://doi.org/10.3390/vaccines9040370
Chicago/Turabian StyleKim, Jihee, Jaewon Yoon, and Jung-Eun Park. 2021. "Construction of Porcine Epidemic Diarrhea Virus-Like Particles and Its Immunogenicity in Mice" Vaccines 9, no. 4: 370. https://doi.org/10.3390/vaccines9040370






