Genetic Analysis Reveals Differences in CD8+ T Cell Epitope Regions That May Impact Cross-Reactivity of Vaccine-Induced T Cells against Wild-Type Mumps Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Set of CD8+ T Cell Epitope Candidates of Mumps Virus (MuV) of Jeryl–Lynn (JL) Vaccine
2.1.1. Generation of Mumps Virus-Infected B-Lymphoblastoid Cell Line
2.1.2. Isolation of Human Leukocyte Antigen Class I-Bound Peptides
2.1.3. Two-Dimensional Reversed-Phase Liquid Chromatography-Mass Spectrometry and Assignment to Mumps Virus Protein Sequences
2.1.4. Previous Described Set of Mumps Virus CD8+ T Cell Epitope Candidates
2.2. Comparison of Amino Acid Sequences of CD8+ T Cell Epitope Candidates with Various Wild-Type MuV Strains
2.3. Prediction of Human Leukocyte Antigen Class-I Binding of T Cell Epitope Candidates of MuV
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rasheed, M.A.U.; Hickman, C.J.; McGrew, M.; Sowers, S.B.; Mercader, S.; Hopkins, A.; Grimes, V.; Yu, T.; Wrammert, J.; Mulligan, M.J.; et al. Decreased humoral immunity to mumps in young adults immunized with MMR vaccine in childhood. Proc. Natl. Acad. Sci. USA 2019, 116, 19071–19076. [Google Scholar] [CrossRef] [Green Version]
- Cardemil, C.V.; Dahl, R.M.; James, L.; Wannemuehler, K.; Gary, H.E.; Shah, M.; Marin, M.; Riley, J.; Feikin, D.R.; Patel, M.; et al. Effectiveness of a Third Dose of MMR Vaccine for Mumps Outbreak Control. N. Engl. J. Med. 2017, 377, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Kaaijk, P.; Wijmenga-Monsuur, A.J.; Van Houten, M.; Veldhuijzen, I.K.; Hulscher, H.I.T.; Kerkhof, J.; Van Der Klis, F.R.; Van Binnendijk, R.S. A Third Dose of Measles-Mumps-Rubella Vaccine to Improve Immunity Against Mumps in Young Adults. J. Infect. Dis. 2019, 221, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Connell, A.R.; Connell, J.; Leahy, T.R.; Hassan, J. Mumps Outbreaks in Vaccinated Populations—Is It Time to Re-assess the Clinical Efficacy of Vaccines? Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Cohen, C.; White, J.M.; Savage, E.J.; Glynn, J.R.; Choi, Y.; Andrews, N.; Brown, D.; Ramsay, M.E. Vaccine effectiveness estimates, 2004–2005 mumps outbreak, England. Emerg. Infect. Dis. 2007, 13, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Rubin, S.A.; Qi, L.; Audet, S.A.; Sullivan, B.; Carbone, K.M.; Bellini, W.J.; Rota, P.A.; Sirota, L.; Beeler, J. Antibody induced by immunization with the Jeryl Lynn mumps vaccine strain effectively neutralizes a heterologous wild-type mumps virus asso-ciated with a large outbreak. J. Infect. Dis. 2008, 198, 508–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayan, G.H.; Rubin, S. Mumps Outbreaks in Vaccinated Populations: Are Available Mumps Vaccines Effective Enough to Prevent Outbreaks? Clin. Infect. Dis. 2008, 47, 1458–1467. [Google Scholar] [CrossRef] [Green Version]
- Gouma, S.; Hulscher, H.I.T.; Klooster, T.M.S.-V.; De Melker, H.E.; Boland, G.J.; Kaaijk, P.; Van Els, C.A.; Koopmans, M.P.; Van Binnendijk, R.S. Mumps-specific cross-neutralization by MMR vaccine-induced antibodies predicts protection against mumps virus infection. Vaccine 2016, 34, 4166–4171. [Google Scholar] [CrossRef]
- Šantak, M.; Lang-Balija, M.; Ivancic-Jelecki, J.; Košutić-Gulija, T.; Ljubin-Sternak, S.; Forčić, D. Antigenic differences between vaccine and circulating wild-type mumps viruses decreases neutralization capacity of vaccine-induced antibodies. Epidemiol. Infect. 2013, 141, 1298–1309. [Google Scholar] [CrossRef]
- Homan, E.J.; Bremel, R.D. Are cases of mumps in vaccinated patients attributable to mismatches in both vaccine T-cell and B-cell epitopes? Hum. Vaccines Immunother. 2013, 10, 290–300. [Google Scholar] [CrossRef] [Green Version]
- Berkhoff, E.G.M.; Geelhoed-Mieras, M.M.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F. Assessment of the extent of variation in influenza A virus cytotoxic T-lymphocyte epitopes by using virus-specific CD8+ T-cell clones. J. Gen. Virol. 2007, 88, 530–535. [Google Scholar] [CrossRef]
- Ueno, T.; Idegami, Y.; Motozono, C.; Oka, S.; Takiguchi, M. Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J. Immunol. 2007, 178, 5513–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, J.; Emmelot, M.; Poelen, M.C.; Van Binnendijk, R.S.; Van Der Lee, S.; Van Baarle, D.; Han, W.G.; Van Els, C.A.; Kaaijk, P. Mumps infection but not childhood vaccination induces persistent polyfunctional CD8 + T-cell memory. J. Allergy Clin. Immunol. 2018, 141, 1908–1911.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jokinen, S.; Österlund, P.; Julkunen, I.; Davidkin, I. Cellular Immunity to Mumps Virus in Young Adults 21 Years after Mea-Sles-Mumps-Rubella Vaccination. J. Infect. Dis. 2007, 196, 861–867. [Google Scholar] [CrossRef]
- Schmidt, M.E.; Varga, S.M. The CD8 T Cell Response to Respiratory Virus Infections. Front. Immunol. 2018, 9, 678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Wit, J.; Emmelot, M.E.; Poelen, M.C.M.; Lanfermeijer, J.; Han, W.G.H.; van Els, C.A.C.M.; Kaaijk, P. The Human CD4 + T Cell Response against Mumps Virus Targets a Broadly Recognized Nucleoprotein Epitope. J. Virol. 2019, 93, e01883-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, J.; Emmelot, M.; Meiring, H.; Brink, J.A.M.V.G.-V.D.; Els, C.A.C.M.V.; Kaaijk, P. Identification of Naturally Processed Mumps Virus Epitopes by Mass Spectrometry: Confirmation of Multiple CD8+ T-Cell Responses in Mumps Patients. J. Infect. Dis. 2019, 221, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Chambers, P.; Rima, B.K.; Duprex, W.P. Molecular differences between two Jeryl Lynn mumps virus vaccine component strains, JL5 and JL2. J. Gen. Virol. 2009, 90, 2973–2981. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Han, W.G.; Emmelot, M.E.; Jaadar, H.; Hulscher, H.I.T.; Van Els, C.A.; Kaaijk, P. Development of an IFNγ ELISPOT for the analysis of the human T cell response against mumps virus. J. Immunol. Methods 2016, 431, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Van Els, C.A.C.M.; Herberts, C.A.; Van Der Heeft, E.; Poelen, M.C.M.; Brink, J.A.M.V.G.-V.D.; Van Der Kooi, A.; Hoogerhout, P.; Hove, G.J.T.; Meiring, H.D.; De Jong, A.P.J.M. A single naturally processed measles virus peptide fully dominates the HLA-A*0201-associated peptide display and is mutated at its anchor position in persistent viral strains. Eur. J. Immunol. 2000, 30, 1172–1181. [Google Scholar] [CrossRef]
- Mei, S.; Ayala, R.; Ramarathinam, S.H.; Illing, P.T.; Faridi, P.; Song, J.; Purcell, A.W.; Croft, N.P. Immunopeptidomic Analysis Reveals That Deamidated HLA-bound Peptides Arise Predominantly from Deglycosylated Precursors. Mol. Cell. Proteom. 2020, 19, 1236–1247. [Google Scholar] [CrossRef]
- Skipper, J.C.; Hendrickson, R.C.; Gulden, P.H.; Brichard, V.; Van Pel, A.; Chen, Y.; Shabanowitz, J.; Wolfel, T.; Slingluff, C.L.; Boon, T.; et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 1996, 183, 527–534. [Google Scholar] [CrossRef]
- Amexis, G.; Rubin, S.; Chizhikov, V.; Pelloquin, F.; Carbone, K.; Chumakov, K. Sequence Diversity of Jeryl Lynn Strain of Mumps Virus: Quantitative Mutant Analysis for Vaccine Quality Control. Virology 2002, 300, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Hill, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Jurtz, V.I.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved Peptide–MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Schellens, I.M.; Meiring, H.D.; Hoof, I.; Spijkers, S.N.; Poelen, M.C.M.; Brink, J.A.M.V.G.-V.D.; Costa, A.I.; Vennema, H.; Kesmir, C.; Van Baarle, D.; et al. Measles Virus Epitope Presentation by HLA: Novel Insights into Epitope Selection, Dominance, and Microvariation. Front. Immunol. 2015, 6, 546. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E. Rates of Molecular Evolution in RNA Viruses: A Quantitative Phylogenetic Analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Kaaijk, P.; Emmelot, M.E.; Meiring, H.; van Els, C.A.C.M.; de Wit, J. Novel Mumps Virus Epitopes Reveal Robust Cytotoxic T Cell Responses After Natural Infection but Not After Vaccination. Sci. Rep. 2021. [Google Scholar] [CrossRef]
- Buseyne, F.; Rivière, Y. The flexibility of the TCR allows recognition of a large set of naturally occurring epitope variants by HIV-specific cytotoxic T lymphocytes. Int. Immunol. 2001, 13, 941–950. [Google Scholar] [CrossRef] [Green Version]
- Rubin, S.; Kennedy, R.; Poland, G. Emerging Mumps Infection. Pediatr. Infect. Dis. J. 2016, 35, 799–801. [Google Scholar] [CrossRef]
- Fiebelkorn, A.P.; Coleman, L.A.; Belongia, E.; Freeman, S.K.; York, D.; Bi, D.; Zhang, C.; Ngo, L.; Rubin, S. Mumps Antibody Response in Young Adults After a Third Dose of Measles-Mumps-Rubella Vaccine. Open Forum Infect. Dis. 2014, 1, ofu094. [Google Scholar] [CrossRef] [PubMed]
- Kaaijk, P.; Nicolaie, M.A.; Van Rooijen, D.; Van Houten, M.; Van Der Klis, F.R.; Buisman, A.-M.; Van Binnendijk, R.S. Dynamics of the antibody response after a third dose of measles-mumps-rubella vaccine indicates a slower decline compared to a second dose. Open Forum Infect. Dis. 2020, 7, ofaa505. [Google Scholar] [CrossRef] [PubMed]
- Almansour, I. Mumps Vaccines: Current Challenges and Future Prospects. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- May, M.; Rieder, C.A.; Rowe, R.J. Emergent lineages of mumps virus suggest the need for a polyvalent vaccine. Int. J. Infect. Dis. 2018, 66, 1–4. [Google Scholar] [CrossRef] [Green Version]
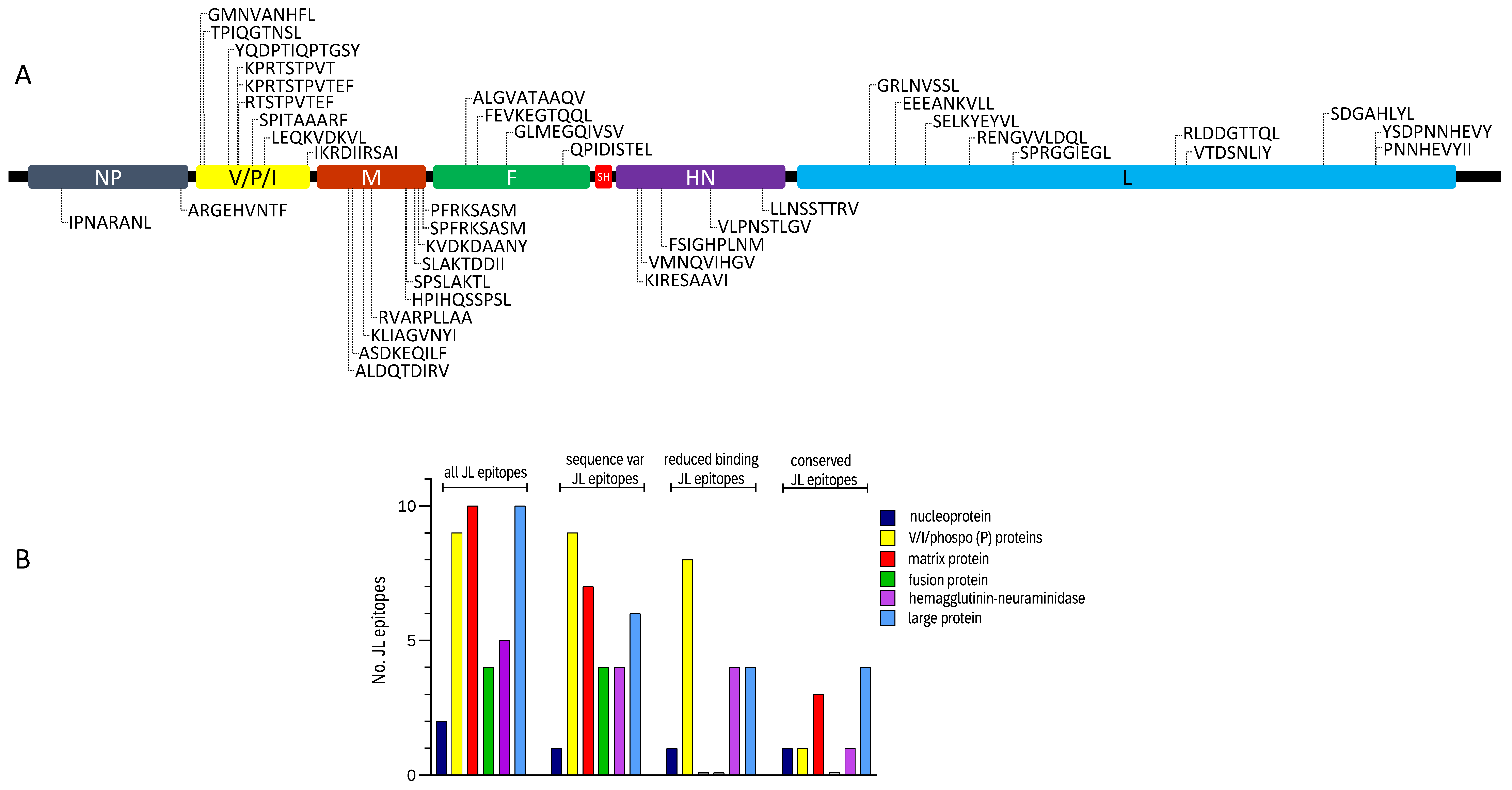
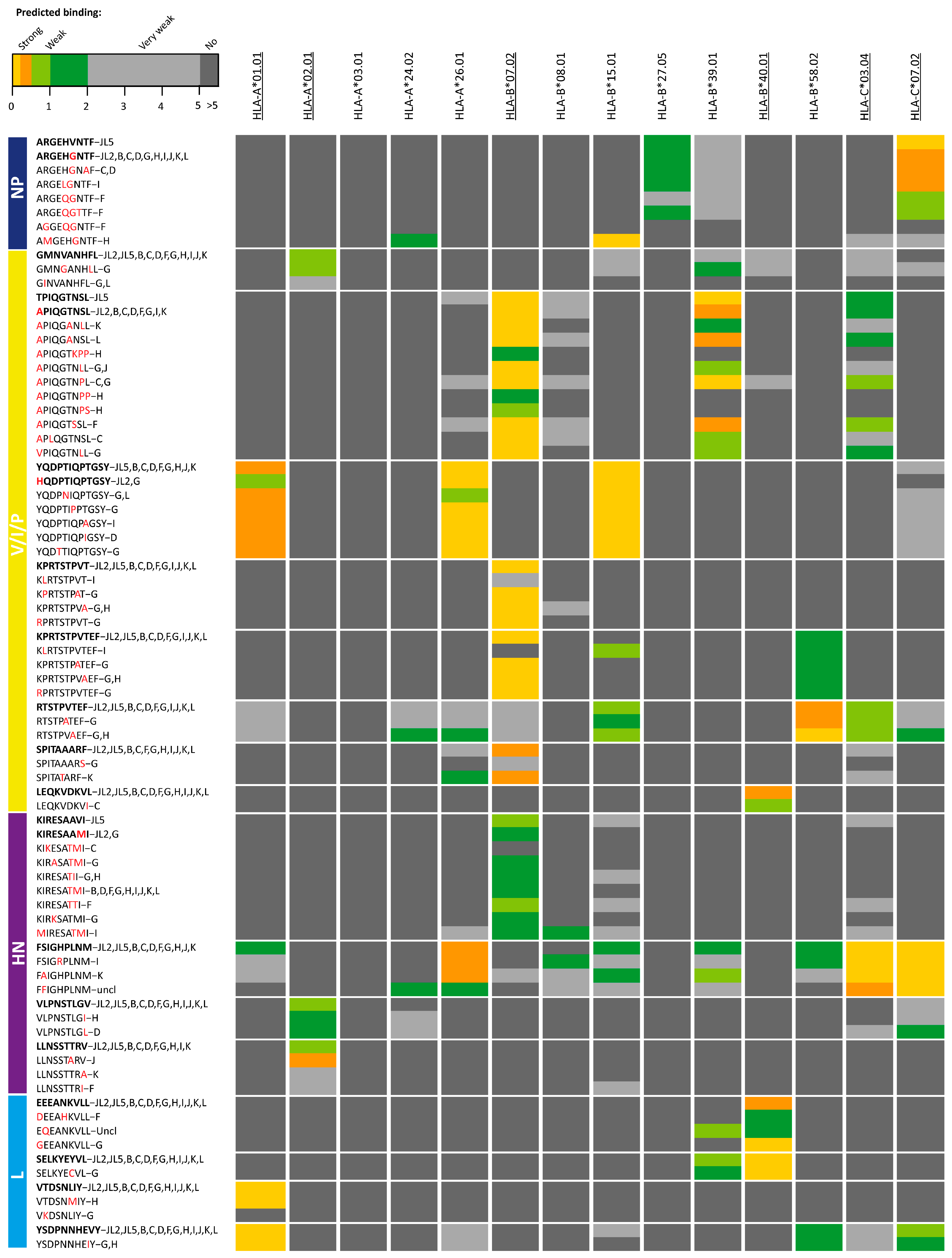
| MuV Protein | Amino Acid Sequence | X-Mer | Protein Location | Described [Ref]/Novel | JL Eluted | Seq Variation | Reduced Binding |
|---|---|---|---|---|---|---|---|
| N | IPNARANL | 8 | 115–122 | [17] | no | no | no |
| N | ARGEHVNTF | 9 | 524–532 | novel | yes | yes | yes |
| V/P/I | GMNVANHFL | 9 | 17–25 | [17] | no | yes | yes |
| V/P/I | TPIQGTNSL | 9 | 27–35 | novel | yes | yes | yes |
| V/P/I | YQDPTIQPTGSY | 12 | 111–122 | [17] | no | yes | yes |
| V/P/I | KPRTSTPVT | 9 | 142–150 | [17] | yes | yes | yes |
| V/P/I | KPRTSTPVTEF | 11 | 142–152 | [17] | yes | yes | yes |
| V/P/I | RTSTPVTEF | 9 | 144–152 | [17] | no | yes | yes |
| V/P/I | SPITAAARF | 9 | 194–202 | [17] | no | yes | yes |
| P | LEQKVDKVL | 9 | 235–243 | novel | yes | yes | yes |
| P | IKRDIIRSAI | 10 | 382–391 | novel | yes | yes | no |
| M | ALDQTDIRV | 9 | 108–116 | [17] | yes | yes | no |
| M | ASDKEQILF | 9 | 120–128 | [17] | no | no | no |
| M | KLIAGVNYI | 9 | 161–169 | [17] | no | yes | no |
| M | RVARPLLAA | 9 | 187–195 | [17] | yes | no | no |
| M | HPIHQSSPSL | 10 | 303–312 | [17] | yes | no | no |
| M | SPSLAKTL | 8 | 309–316 | [17] | yes | yes | no |
| M | SLAKTDDII | 9 | 337–345 | [17] | no | yes | no |
| M | KVDKDAANY | 9 | 350–358 | [17] | no | yes | no |
| M | SPFRKSASM | 9 | 364–372 | [17] | yes | yes | no |
| M | PFRKSASM | 8 | 365–372 | [17] | no | yes | no |
| F | ALGVATAAQV | 10 | 112–121 | [17] | yes | yes | no |
| F | FEVKEGTQQL | 10 | 152–161 | [17] | no | yes | no |
| F | GLMEGQIVSV | 10 | 253–262 | [17] | yes | yes | no |
| F | QPIDISTEL | 9 | 445–453 | [17] | no | yes | no |
| HN | KIRESAAVI | 9 | 74–82 | novel | yes | yes | yes |
| HN | VMNQVIHGV | 9 | 88–96 | [17] | yes | no | no |
| HN | FSIGHPLNM | 9 | 157–165 | [17] | no | yes | yes |
| HN | VLPNSTLGV 1 | 9 | 326–334 | [17] | no | yes | yes |
| HN | LLNSSTTRV 1 | 9 | 505–513 | [17] | no | yes | yes |
| L | GRLNVSSL 1 | 8 | 249–256 | [17] | yes | no | no |
| L | EEEANKVLL | 9 | 336–344 | [17] | no | yes | yes |
| L | SELKYEYVL | 9 | 441–449 | [17] | no | yes | yes |
| L | RENGVVLDQL | 10 | 591–600 | [17] | no | no | no |
| L | SPRGGIEGL | 9 | 740–748 | [17] | yes | no | no |
| L | RLDDGTTQL | 9 | 1299–1307 | [17] | no | no | no |
| L | VTDSNLIY | 8 | 1336–1343 | [17] | no | yes | yes |
| L | SDGAHLYL | 8 | 1806–1813 | [17] | yes | yes | no |
| L | YSDPNNHEVY 1 | 10 | 1983–1992 | [17] | yes | yes | yes |
| L | PNNHEVYII 1 | 9 | 1986–1994 | novel | yes | yes | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaaijk, P.; Emmelot, M.E.; Kerkhof, J.; van Els, C.A.C.M.; Meiring, H.D.; de Wit, J.; Bodewes, R. Genetic Analysis Reveals Differences in CD8+ T Cell Epitope Regions That May Impact Cross-Reactivity of Vaccine-Induced T Cells against Wild-Type Mumps Viruses. Vaccines 2021, 9, 699. https://doi.org/10.3390/vaccines9070699
Kaaijk P, Emmelot ME, Kerkhof J, van Els CACM, Meiring HD, de Wit J, Bodewes R. Genetic Analysis Reveals Differences in CD8+ T Cell Epitope Regions That May Impact Cross-Reactivity of Vaccine-Induced T Cells against Wild-Type Mumps Viruses. Vaccines. 2021; 9(7):699. https://doi.org/10.3390/vaccines9070699
Chicago/Turabian StyleKaaijk, Patricia, Maarten E. Emmelot, Jeroen Kerkhof, Cécile A.C.M. van Els, Hugo D. Meiring, Jelle de Wit, and Rogier Bodewes. 2021. "Genetic Analysis Reveals Differences in CD8+ T Cell Epitope Regions That May Impact Cross-Reactivity of Vaccine-Induced T Cells against Wild-Type Mumps Viruses" Vaccines 9, no. 7: 699. https://doi.org/10.3390/vaccines9070699
APA StyleKaaijk, P., Emmelot, M. E., Kerkhof, J., van Els, C. A. C. M., Meiring, H. D., de Wit, J., & Bodewes, R. (2021). Genetic Analysis Reveals Differences in CD8+ T Cell Epitope Regions That May Impact Cross-Reactivity of Vaccine-Induced T Cells against Wild-Type Mumps Viruses. Vaccines, 9(7), 699. https://doi.org/10.3390/vaccines9070699






