VENUS, a Novel Selection Approach to Improve the Accuracy of Neoantigens’ Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of NGS Data from Patients with Solid Tumours
- -
- mutation allele frequency (MF) in the tumor DNA sample ≥ 10%;
- -
- ratio of the MF in the tumor DNA sample and in the control DNA sample ≥ 5;
- -
- number of mutated reads at chromosomal position of somatic variant in the tumor DNA > 2;
- -
- number of mutated reads at chromosomal position of somatic variant in the normal DNA < 2.
2.2. Exome and RNA Sequencing of Mice Tumors
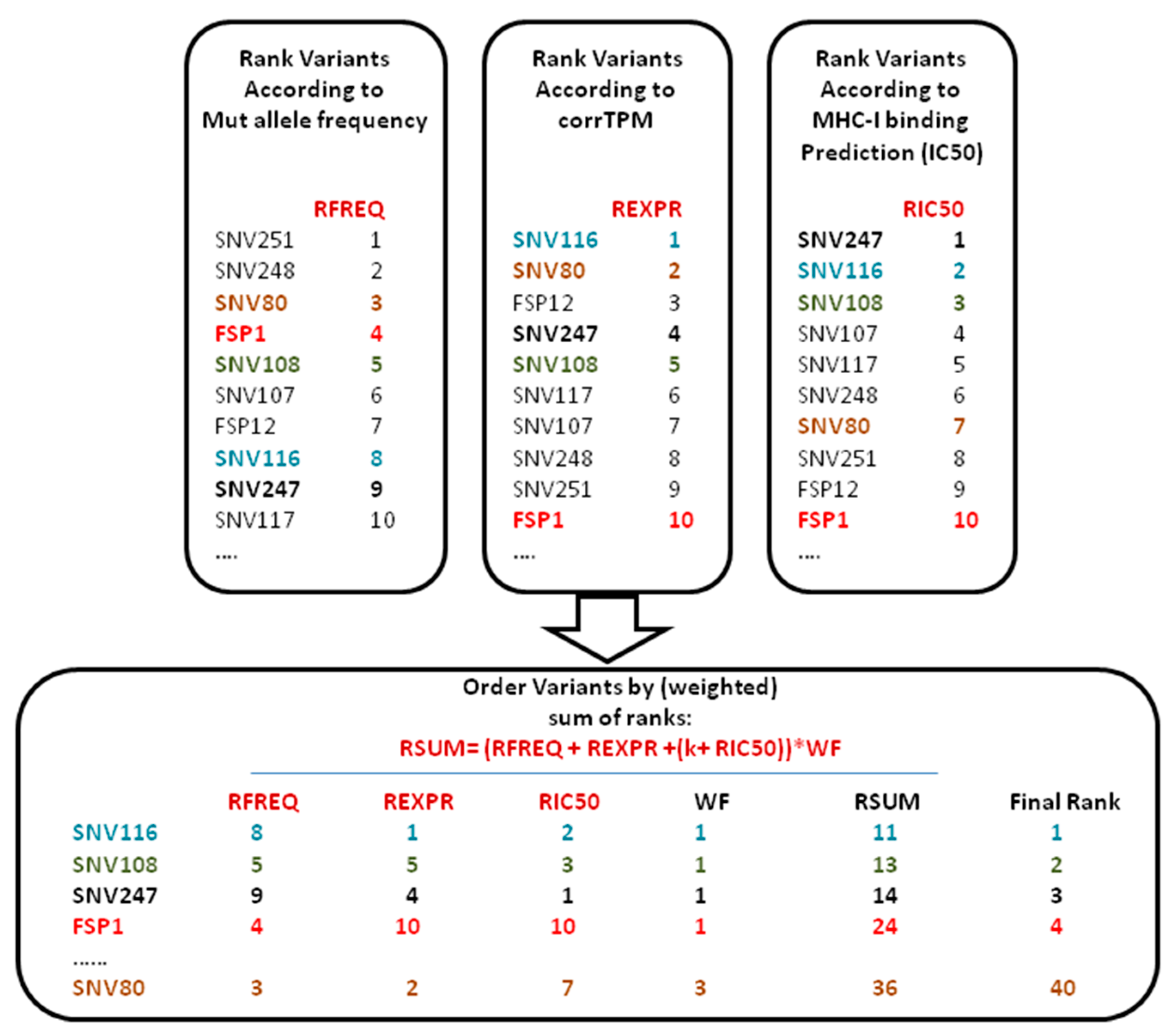
2.3. VENUS RSUM Score
2.4. GAd Vector Production
2.5. Mice
2.6. In Vivo Tumor Growth
2.7. In Vivo Treatments
2.8. Ex Vivo Immune Analysis
3. Results
3.1. VENUS RSUM Score Provides a Ranked List of Neo-Peptides Ordered According to a Balanced Three-Parameter Score
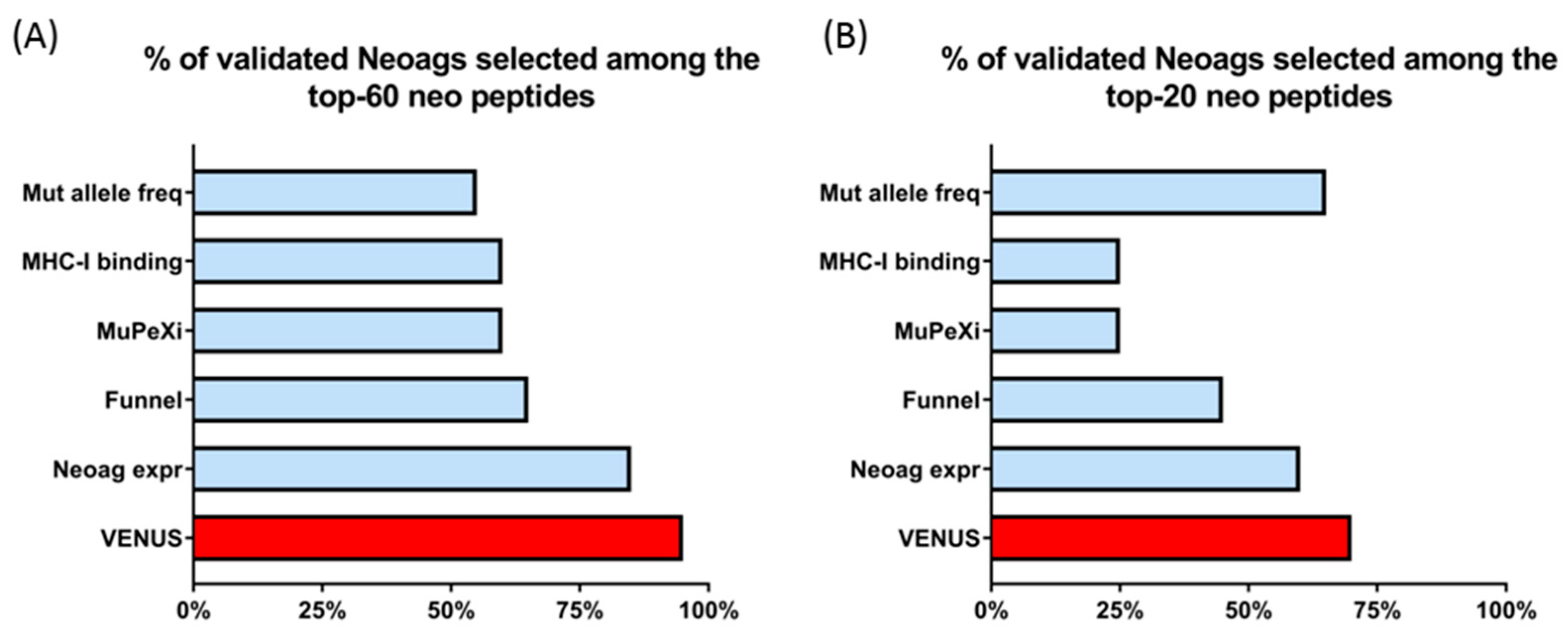
3.2. Venus RSUM Score Captures Validated Neoantigens Eliciting an Immune Response in Humans
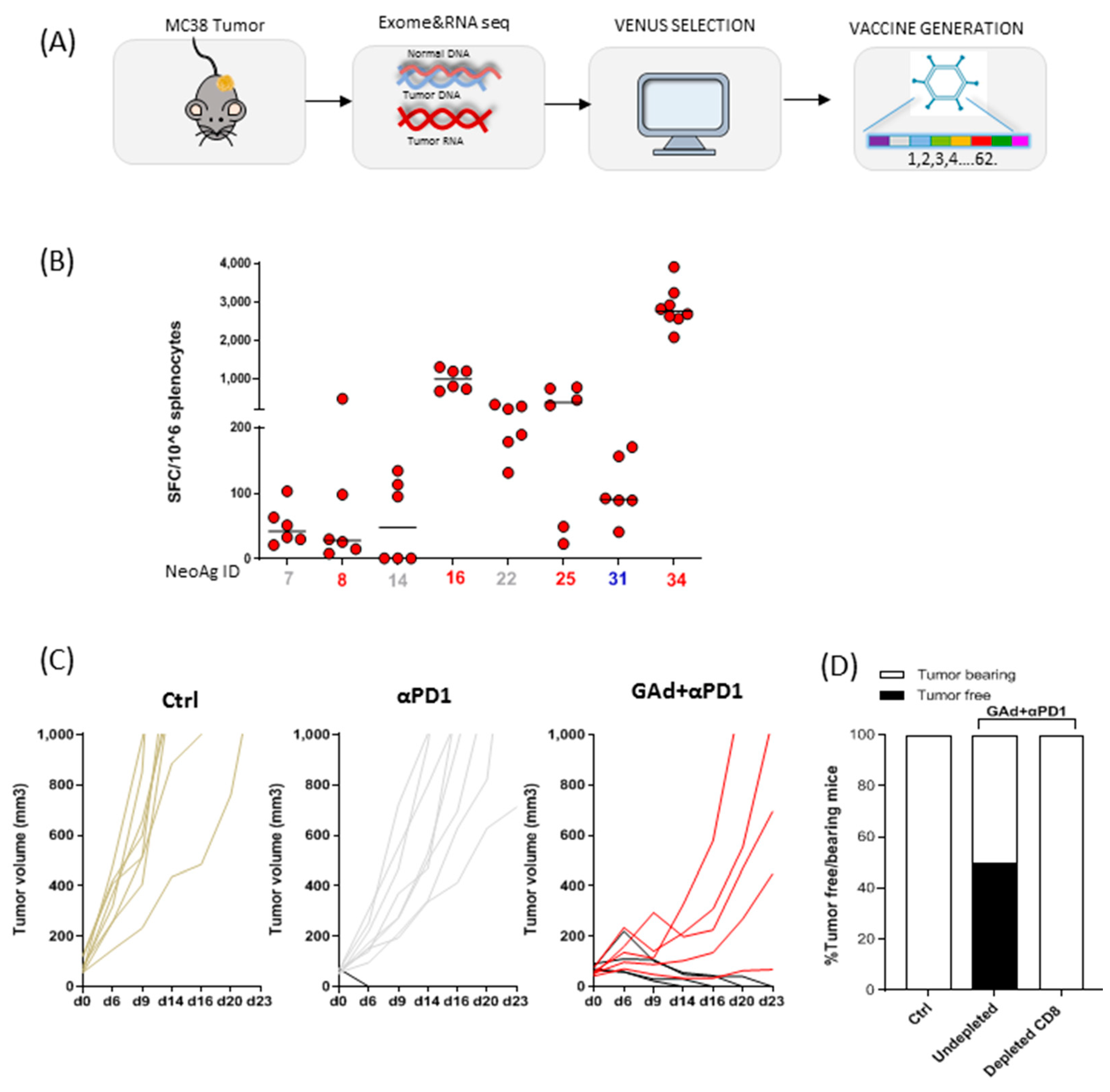
3.3. Generation of Viral Vectors Targeting the Best Neoantigens Ranked by VENUS in MC38 Tumour Model
3.4. Vaccination with VENUS-Identified Neoantigens Is Effective in Eradicating Large MC38 Tumours in Combination with Anti-PD1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CPIs | Check point inhibitors |
| NGS | Next generation sequencing |
| SNVs | Single nucleotide variants |
| FSP | Frame shift peptide |
| MHC | Major histocompatibility complex |
| VENUS | Vaccine-encoded neoantigens unrestricted selection |
| MF | Mutation allele frequency |
| TPM | Transcripts for million |
| corrTPM | corrected transcripts for million |
| TPA | Tissue plasminogen activator |
| TetO | Tet operator |
| Ad | Adenoviral vector |
| GAd | Great apes-derived adenovirus |
| TIL | Tumor infiltrating lymphocytes |
| s.c | Subcutaneous |
| i.v | Intravenous |
| i.m | Intramuscle |
References
- Gajewski, T.F.; Meng, Y.; Harlin, H. Immune suppression in the tumor microenvironment. J. Immunother. 2006, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Shuqiang, L.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.-P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, F.; Xin, K.; Wang, Q.; Yu, L.; Liu, B.; Liu, Q. Cancer-Testis Antigen Peptide Vaccine for Cancer Immunotherapy: Progress and Prospects. Transl. Oncol. 2019, 12, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.G.; Li, F.; Roszik, J.; Lizee, G. Exploiting Tumor Neoantigens to Target Cancer Evolution: Current Challenges and Promising Therapeutic Approaches. Cancer Discov. 2021, 11, 1024–1039. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Turajlic, S.; Litchfield, K.; Xu, H.; Rosenthal, R.; McGranahan, N.; Reading, J.; Wong, Y.N.S.; Rowan, A.; Kanu, N.; Al Bakir, M.; et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 2017, 18, 1009–1021. [Google Scholar] [CrossRef] [Green Version]
- Łuksza, M.; Riaz, N.; Makarov, V.; Balachandran, V.P.; Hellmann, M.D.; Solovyov, A.; Rizvi, N.A.; Merghoub, T.; Levine, A.J.; Chan, T.A.; et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017, 551, 517–520. [Google Scholar] [CrossRef]
- Wells, D.K.; van Buuren, M.M.; Dang, K.K.; Hubbard-Lucey, V.M.; Sheehan, K.C.; Campbell, K.M.; Lamb, A.; Ward, J.P.; Sidney, J.; Blazquez, A.B.; et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell 2020, 183, 818–834.e13. [Google Scholar] [CrossRef]
- Gros, A.; Parkhurst, M.R.; Tran, E.; Pasetto, A.; Robbins, P.F.; Ilyas, S.; Pirckett, T.D.; Gartner, J.J.; Crystal, J.S.; Roberts, I.M.; et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 2016, 22, 433–438. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.B.; Daly, M.J.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Bergmann, A.E.; Arora, K.; Vacic, V.; Zody, M.C.; Iossifov, I.; O’Rawe, A.J.; Wu, Y.; Barron, L.T.J.; Rosenbaum, J.; et al. Indel variant analysis of short-read sequencing data with Scalpel. Nat. Protoc. 2016, 11, 2529–2548. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, O.M.; Whitwham, A.; Keane, T.; McCarthy, A.S.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Szolek, A.; Schubert, B.; Mohr, C.; Sturm, M.; Feldhahn, M.; Kohlbacher, O. OptiType: Precision HLA typing from next-generation sequencing data. Bioinformatics 2014, 30, 3310–3316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moutaftsi, M.; Peters, B.; Pasquetto, V.; Tscharke, D.C.; Sidney, J.; Bui, H.H.; Grey, H.; Sette, A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006, 24, 817–819. [Google Scholar] [CrossRef]
- D’Alise, A.M.; Leoni, G.; Cotugno, G.; Troise, F.; Langone, F.; Fichera, I.; De Lucia, M.; Avalle, L.; Vitale, R.; Leuzzi, A.; et al. Adenoviral vaccine targeting multiple neoantigens as strategy to eradicate large tumors combined with checkpoint blockade. Nat. Commun. 2019, 10, 2688. [Google Scholar] [CrossRef] [Green Version]
- Gros, A.; Tran, E.; Parkhurst, M.R.; Ilyas, S.; Pasetto, A.; Groh, E.M.; Robbins, P.F.; Yossef, R.; Garcia-Garijo, A.; Fajardo, C.A.; et al. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J. Clin. Investig. 2019, 129, 4992–5004. [Google Scholar] [CrossRef] [Green Version]
- Tran, E.; Ahmadzadeh, M.; Lu, Y.C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Bjerregaard, A.M.; Nielsen, M.; Hadrup, S.R.; Szallasi, Z.; Eklund, A.C. MuPeXI: Prediction of neo-epitopes from tumor sequencing data. Cancer Immunol. Immunother. 2017, 66, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.T.; Lupardus, P.J.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, J.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Bassani-Sternberg, M.; Pletscher-Frankild, S.; Jensen, L.J.; Mann, M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteomics 2015, 14, 658–673. [Google Scholar] [CrossRef] [Green Version]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonsack, M.; Hoppe, S.; Winter, J.; Tichy, D.; Zeller, C.; Kupper, M.D.; Schitter, E.C.; Blatnik, R.; Riemer, A.B. Performance Evaluation of MHC Class-I Binding Prediction Tools Based on an Experimentally Validated MHC-Peptide Binding Data Set. Cancer Immunol. Res. 2019, 7, 719–736. [Google Scholar] [CrossRef]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Borthwick, N.; Ahmed, T.; Ondondo, B.; Hayes, P.; Rose, A.; Ebrahimsa, U.; Hayton, E.-J.; Black, A.; Bridgeman, A.; Rosario, M.; et al. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol. Ther. 2014, 22, 464–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, E.; Folgori, A.; Capone, S.; Swadling, L.; Aston, S.; Kurioka, A.; Meyer, J.; Huddart, R.; Smith, K.; Townsend, R.; et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci. Transl. Med. 2012, 4, 115ra1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassani-Sternberg, M.; Bräunlein, E.; Klar, R.; Engleitner, T.; Sinitcyn, P.; Audehm, S.; Straub, M.; Weber, J.; Slotta-Huspenina, J.; Specht, K.; et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 2016, 7, 13404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
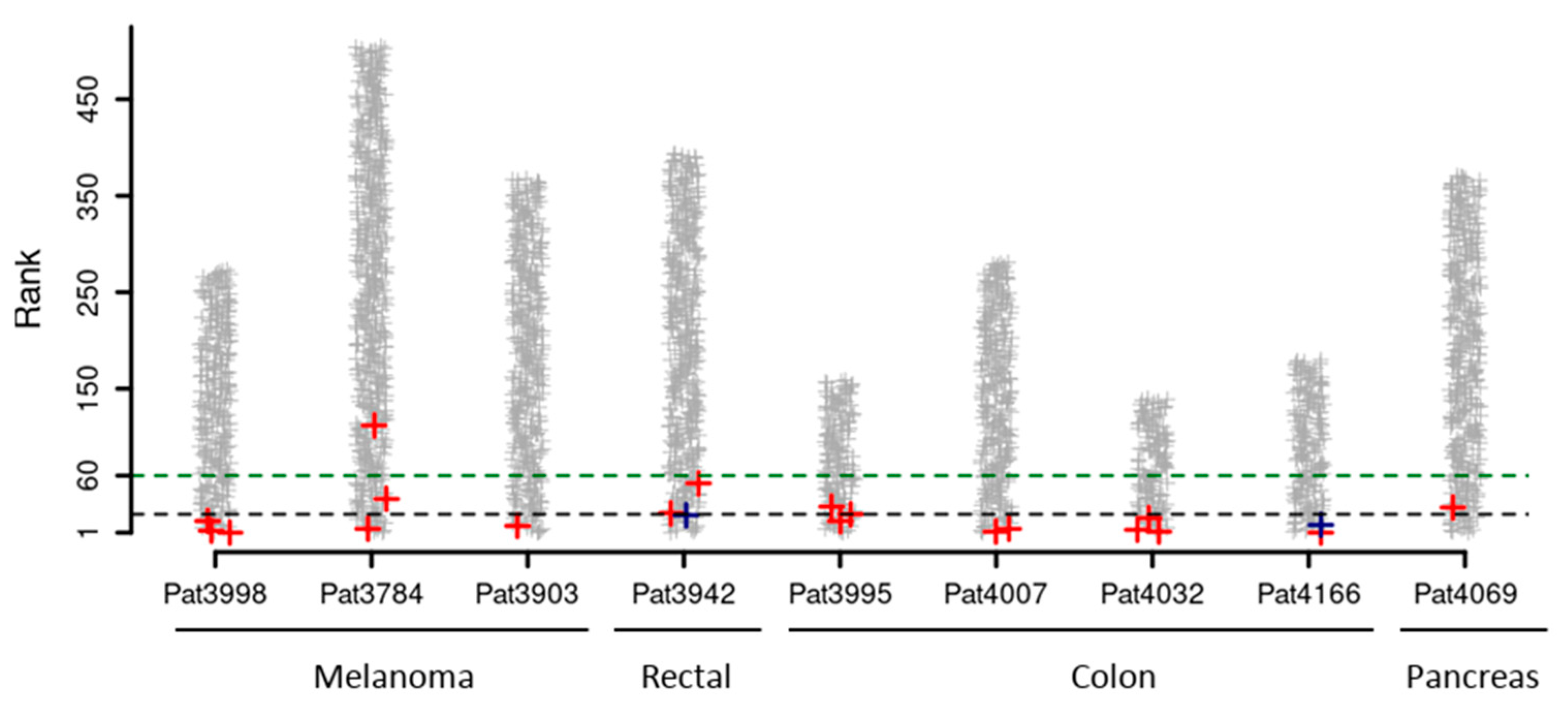
| Tumor Type | Patient ID | Total Detected Neo-Peptides | Experimentally Validated Neoantigens | Position in VENUS Ranked List | Percentage of Neo-Peptides Ranked Better |
|---|---|---|---|---|---|
| Melanoma | 3998 | 268 | MAGEA6_E168K | 1 | 0% |
| PDS5A_H1007Y | 3 | 1% | |||
| MED13_P1691S | 13 | 5% | |||
| 3784 | 494 | FLNA_R2049C | 5 | 1% | |
| SON_R1927C | 36 | 7% | |||
| KIF16B_L1009P | 110 | 22% | |||
| 3903 | 435 | KIF1BP_P246S | 8 | 2% | |
| Rectal | 3942 | 396 | GPD2_E426K | 19 | 5% |
| NUP98_A359D | 21 | 5% | |||
| KARS_D328H | 58 | 15% | |||
| Colon | 3995 | 138 | RNF213_N1702S | 13 | 9% |
| TUBGCP2_P265L | 20 | 14% | |||
| KRAS_G12D | 28 | 20% | |||
| 4007 | 262 | SKIV2L_R653H | 1 | 0% | |
| 4032 | 136 | API5_R243Q | 2 | 1% | |
| PHLPP1_G566E | 4 | 3% | |||
| RNF10_E572K | 16 | 12% | |||
| 4166 | 180 | NPLOC_G1473V | 1 | 0% | |
| SUN1_A127T | 9 | 5% | |||
| Pancreas | 4069 | 371 | ZFYVE27_R6H | 41 | 11% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leoni, G.; D’Alise, A.M.; Tucci, F.G.; Micarelli, E.; Garzia, I.; De Lucia, M.; Langone, F.; Nocchi, L.; Cotugno, G.; Bartolomeo, R.; et al. VENUS, a Novel Selection Approach to Improve the Accuracy of Neoantigens’ Prediction. Vaccines 2021, 9, 880. https://doi.org/10.3390/vaccines9080880
Leoni G, D’Alise AM, Tucci FG, Micarelli E, Garzia I, De Lucia M, Langone F, Nocchi L, Cotugno G, Bartolomeo R, et al. VENUS, a Novel Selection Approach to Improve the Accuracy of Neoantigens’ Prediction. Vaccines. 2021; 9(8):880. https://doi.org/10.3390/vaccines9080880
Chicago/Turabian StyleLeoni, Guido, Anna Morena D’Alise, Fabio Giovanni Tucci, Elisa Micarelli, Irene Garzia, Maria De Lucia, Francesca Langone, Linda Nocchi, Gabriella Cotugno, Rosa Bartolomeo, and et al. 2021. "VENUS, a Novel Selection Approach to Improve the Accuracy of Neoantigens’ Prediction" Vaccines 9, no. 8: 880. https://doi.org/10.3390/vaccines9080880
APA StyleLeoni, G., D’Alise, A. M., Tucci, F. G., Micarelli, E., Garzia, I., De Lucia, M., Langone, F., Nocchi, L., Cotugno, G., Bartolomeo, R., Romano, G., Allocca, S., Troise, F., Nicosia, A., Lahm, A., & Scarselli, E. (2021). VENUS, a Novel Selection Approach to Improve the Accuracy of Neoantigens’ Prediction. Vaccines, 9(8), 880. https://doi.org/10.3390/vaccines9080880





