Recombinant Antigen of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-2) Promotes M1 Repolarization of Porcine Alveolar Macrophages and Th1 Type Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Pigs and Inoculations
2.3. Constructed the Recombinant Protein Antigen
2.4. Collecting Porcine Alveolar Macrophages (PAMs)
2.5. Cytokine Stimulation
2.6. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
2.7. Next Generation Sequencing (NGS) Analysis
2.8. Integration of the Protein-Protein Interaction (PPI) Network
2.9. Isolation of Porcine Peripheral Blood Mononuclear Cells (PBMC)
2.10. Fluorescence Activated Cell Sorting (FACS) for T-Cell Subsets
2.11. Flow Cytometry Analysis
2.12. Th1 Cytokines Assay
2.13. Statistical Analysis
3. Results
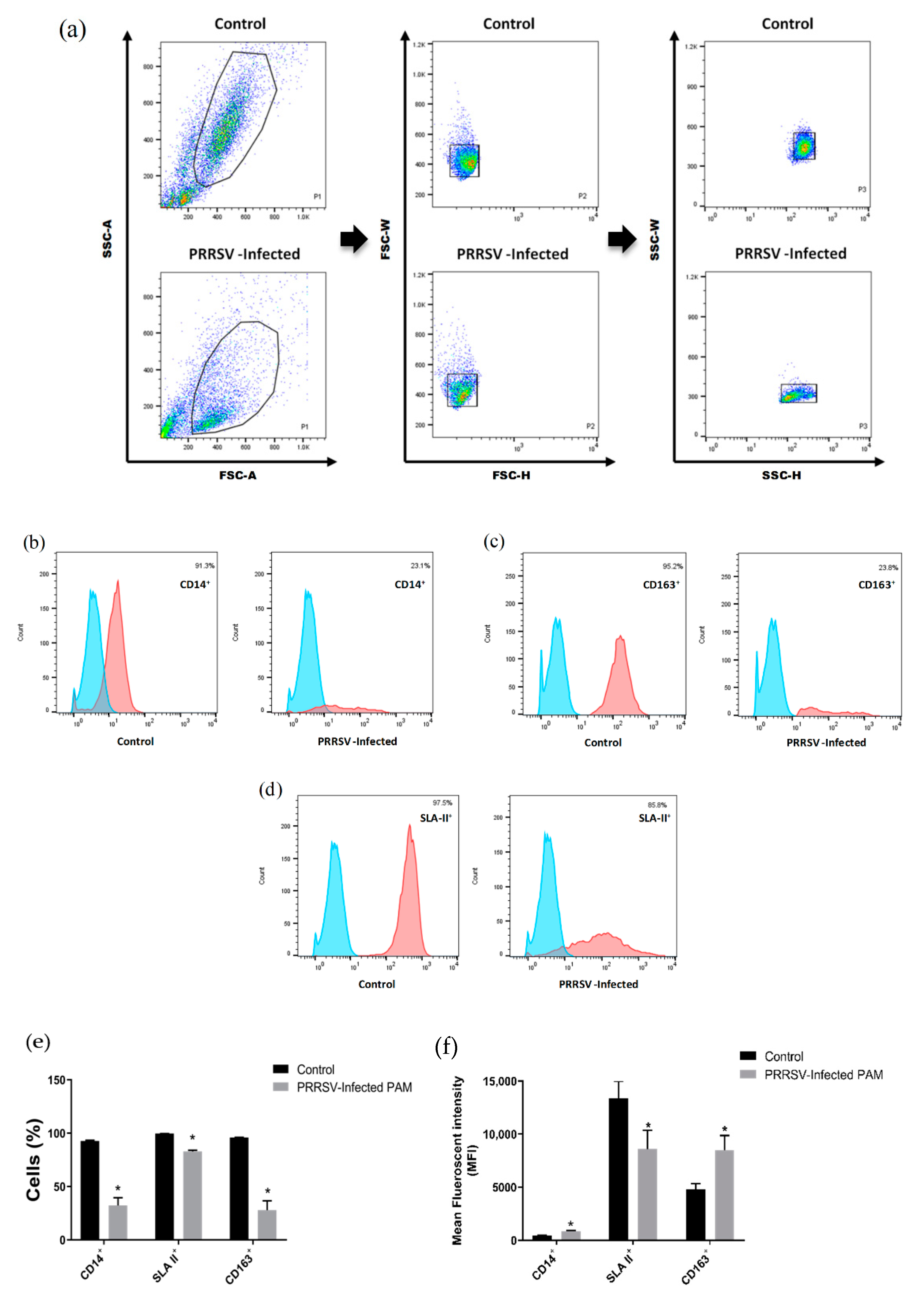
3.1. A1 Directs Macrophages Polarization toward M1 Macrophages and Downregulation of CD163 Expression
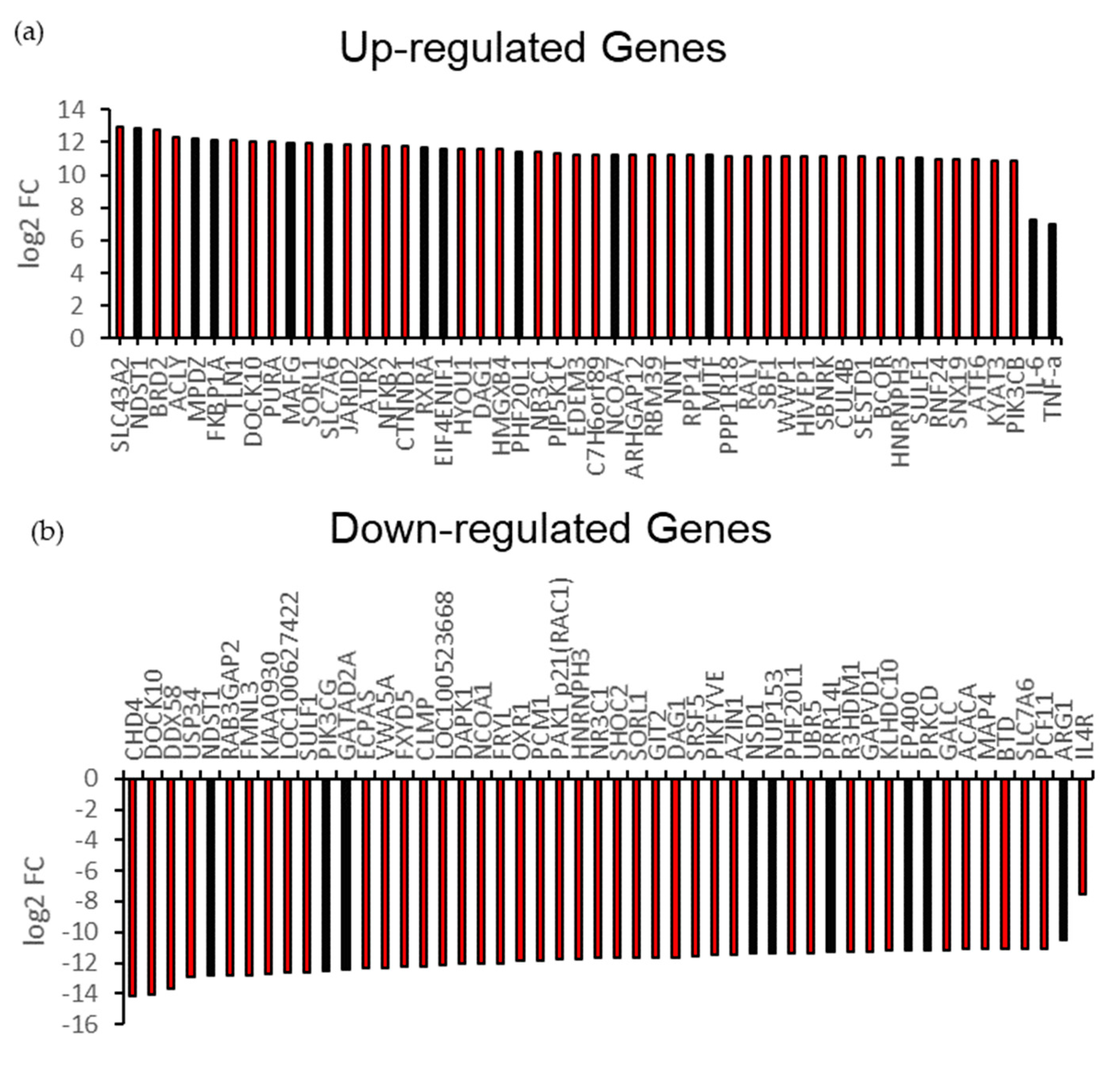
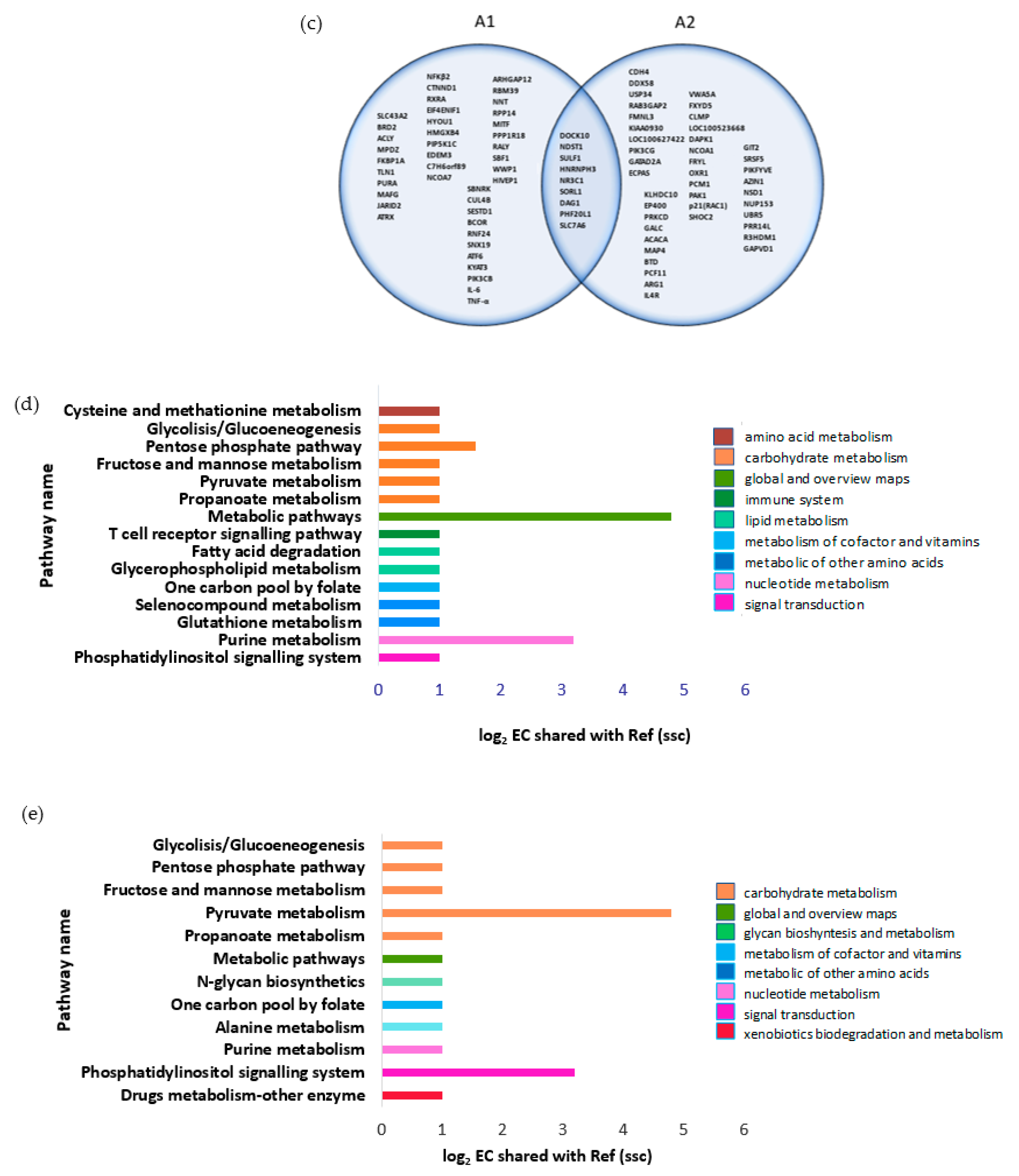
3.2. A1 Stimulate Endogenous Pro-Inflammatory Gene for T-Cell Receptor (TCR) Signaling Pathway
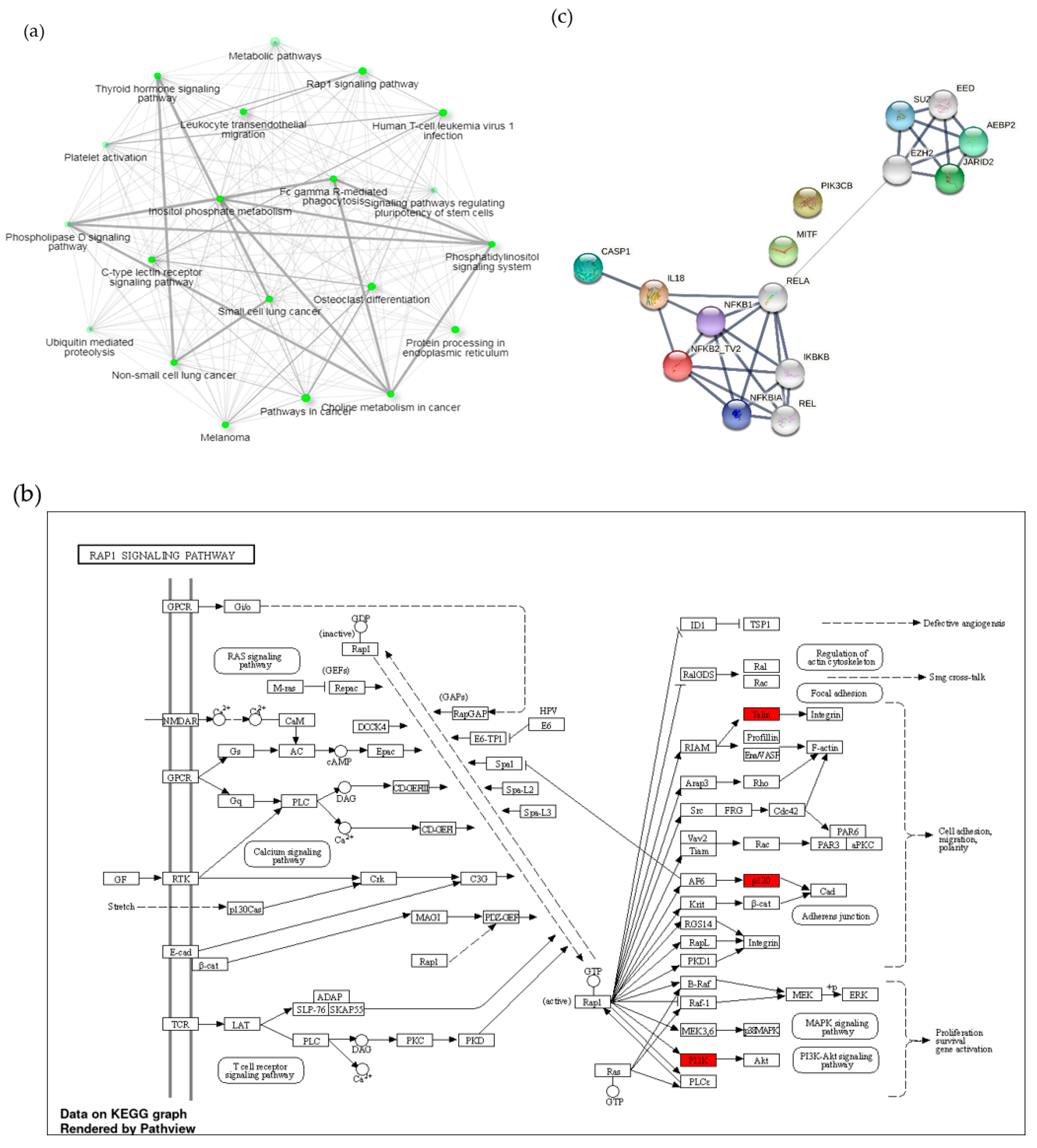
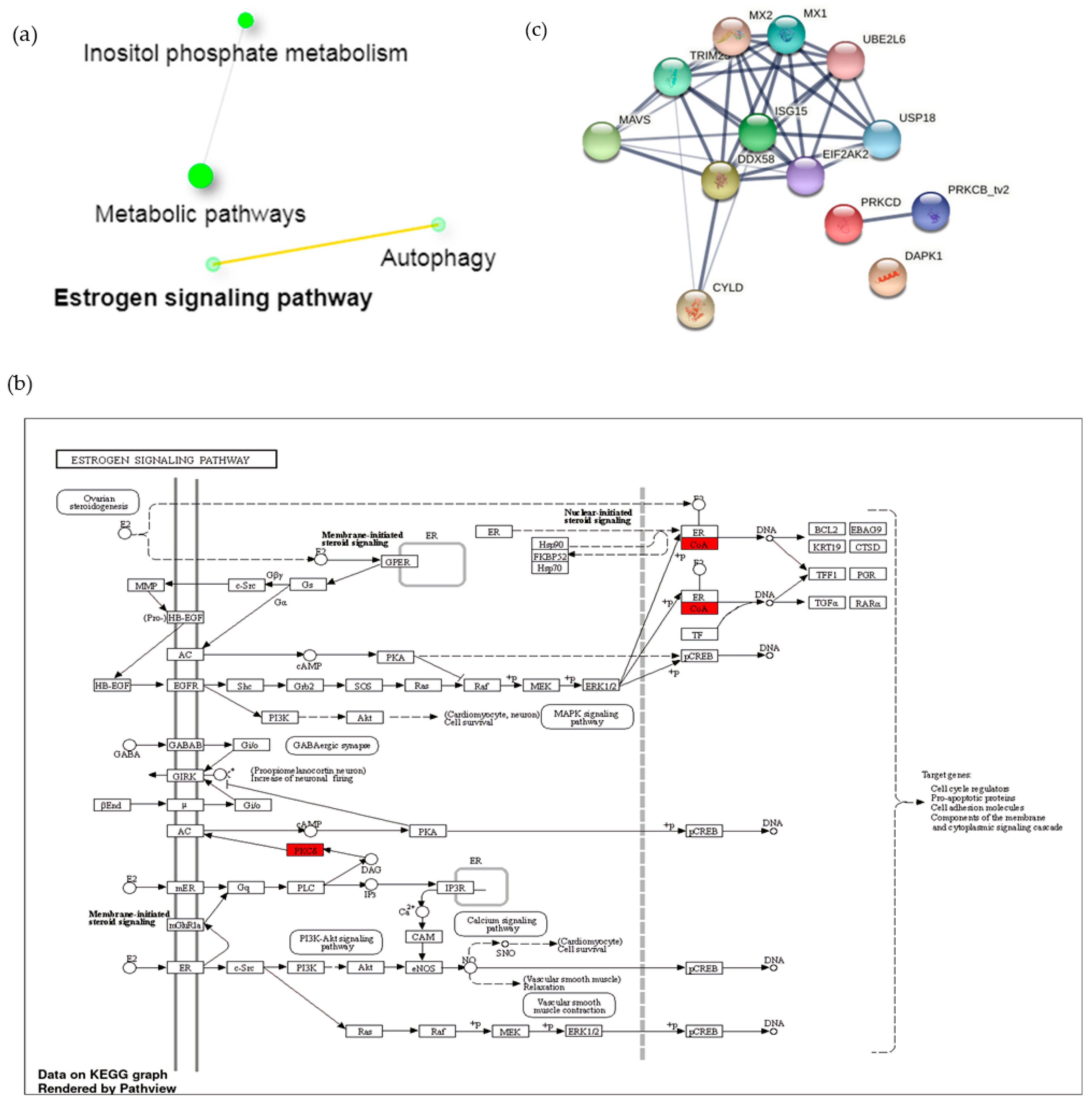
3.3. A1 Potentially Regulate Immune Response by Promoting Rap1 Signaling Pathway and Protein-Protein Interaction (PPI) Network
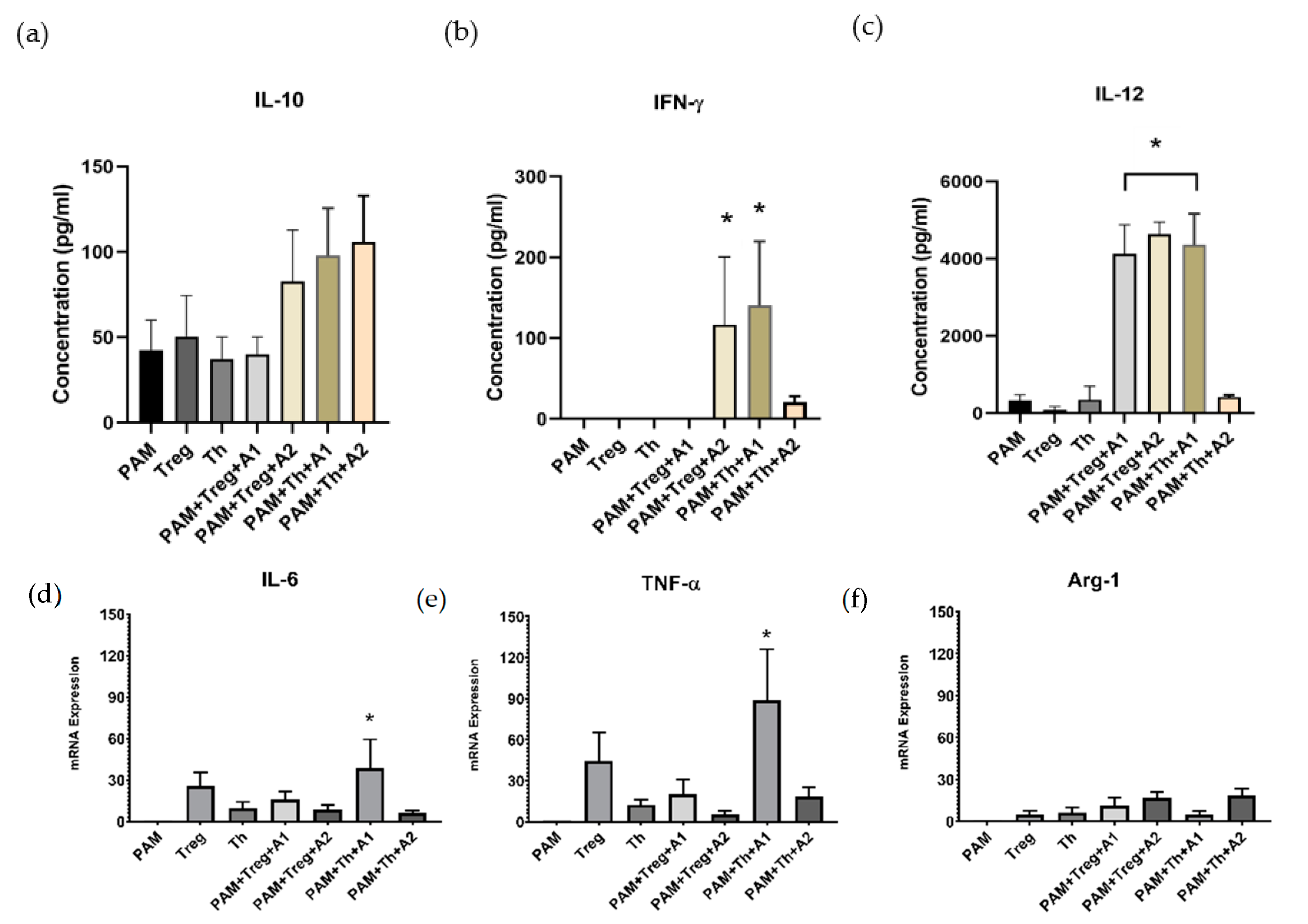
3.4. PAMs Induced by A1 Activate Th1-Cells by Boosting Th1 Cytokine Secretion and TNF-α Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinton, M.; Gulyaeva, A.; Balasuriya, U.; Dunowska, M.; Faaberg, K.S.; Goldberg, T.; Leung, F.C.C.; Nauwynck, H.J.; Snijder, E.J.; Stadejek, T.; et al. ICTV Virus Taxonomy Profile: Arteriviridae. J. Gen. Virol. 2021, 102, 001632. [Google Scholar] [CrossRef]
- Li, X.; Galliher-Beckley, A.; Pappan, L.; Trible, B.; Kerrigan, M.; Beck, A.; Hesse, R.; Blecha, F.; Nietfeld, J.C.; Rowland, R.R.; et al. Comparison of host immune responses to homologous and heterologous type II porcine reproductive and respiratory syndrome virus (PRRSV) challenge in vaccinated and unvaccinated pigs. BioMed Res. Int. 2014, 2014, 416727. [Google Scholar] [CrossRef]
- Wissink, E.H.; Kroese, M.V.; Van Wijk, H.A.; Rijsewijk, F.A.; Meulenberg, J.J.; Rottier, P.J. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 2005, 79, 12495–12506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durán, M.G.; Costa, S.; Sarraseca, J.; de la Roja, N.; García, J.; García, I.; Rodríguez, M.J. Generation of porcine reproductive and respiratory syndrome (PRRS) virus-like-particles (VLPs) with different protein composition. J. Virol. Methods 2016, 236, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Pesente, P.; Rebonato, V.; Sandri, G.; Giovanardi, D.; Ruffoni, L.S.; Torriani, S. Phylogenetic analysis of ORF5 and ORF7 sequences of porcine reproductive and respiratory syndrome virus (PRRSV) from PRRS-positive Italian farms: A showcase for PRRSV epidemiology and its consequences on farm management. Vet. Microbiol. 2006, 114, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Yu, J.E.; Shin, J.E.; Kang, A.; Kim, W.I.; Lee, C.; Lee, J.; Cho, I.S.; Choe, S.E.; Cha, S.H. Geographic distribution and molecular analysis of porcine reproductive and respiratory syndrome viruses circulating in swine farms in the Republic of Korea between 2013 and 2016. BMC Vet. Res. 2018, 14, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular innate immunity against PRRSV and swine influenza viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, P.B.; Dinh, P.X.; Ansari, I.H.; De Lima, M.; Osorio, F.A.; Pattnaik, A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010, 84, 1731–1740. [Google Scholar] [CrossRef] [Green Version]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479, 475–486. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Yoo, D. PRRS virus receptors and their role for pathogenesis. Vet. Microbiol. 2015, 177, 229–241. [Google Scholar] [CrossRef]
- Fang, L.; Jiang, Y.; Xiao, S.; Niu, C.; Zhang, H.; Chen, H. Enhanced immunogenicity of the modified GP5 of porcine reproductive and respiratory syndrome virus. Virus Genes 2006, 32, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Jiang, P.; Li, Y.; Zeshan, B.; Cao, J.; Wang, X. GM-CSF fused with GP3 and GP5 of porcine reproductive and respiratory syndrome virus increased the immune responses and protective efficacy against virulent PRRSV challenge. Virus Res. 2009, 143, 24–32. [Google Scholar] [CrossRef]
- Lewinsohn, D.A.; Lewinsohn, D.M.; Scriba, T.J. Polyfunctional CD4+ T-cells as targets for tuberculosis vaccination. Front. Immunol. 2017, 8, 1262. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Dai, A.; Fan, J.; Li, Y.; Chen, A.; Zhou, X.; Luo, M.; Yang, X.; Liu, J. Efficacy of Type 2 PRRSV vaccine against challenge with the Chinese lineage 1 (NADC30-like) PRRSVs in pigs. Sci. Rep. 2019, 9, 10781. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: Implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000, 74, 309–329. [Google Scholar] [CrossRef]
- Mengeling, W.L.; Lager, K.M.; Vorwald, A.C.; Koehler, K.J. Strain specificity of the immune response of pigs following vaccination with various strains of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2003, 93, 13–24. [Google Scholar] [CrossRef]
- Opriessnig, T.; Halbur, P.G.; Yoon, K.J.; Pogranichniy, R.M.; Harmon, K.M.; Evans, R.; Key, K.F.; Pallares, F.J.; Thomas, P.; Meng, X.J. Comparison of molecular and biological characteristics of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (ingelvac PRRS MLV), the parent strain of the vaccine (ATCC VR2332), ATCC VR2385, and two recent field isolates of PRRSV. J. Virol. 2002, 76, 11837–11844. [Google Scholar]
- Nascimento, I.P.; Leite, L.C. Recombinant vaccines and the development of new vaccine strategies. Braz. J. Med. Biol. Res. 2012, 45, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Clem, A.S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011, 3, 73. [Google Scholar] [CrossRef] [PubMed]
- Salerno-Gonçalves, R.; Sztein, M.B. Cell-mediated immunity and the challenges for vaccine development. Trends Microbiol. 2006, 14, 536–542. [Google Scholar] [CrossRef]
- Wang, T.Y.; Liu, Y.G.; Li, L.; Wang, G.; Wang, H.M.; Zhang, H.L.; Zhao, S.F.; Gao, J.C.; An, T.Q.; Tian, Z.J.; et al. Porcine alveolar macrophage CD163 abundance is a pivotal switch for porcine reproductive and respiratory syndrome virus infection. Oncotarget 2018, 9, 12174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezquerra, A.; Revilla, C.; Alvarez, B.; Perez, C.; Alonso, F.; Dominguez, J. Porcine myelomonocytic markers and cell populations. Dev. Comp. Immunol. 2009, 33, 284–298. [Google Scholar] [CrossRef]
- Calvert, J.G.; Slade, D.E.; Shields, S.L.; Jolie, R.; Mannan, R.M.; Ankenbauer, R.G.; Welch, S.K.W. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007, 81, 7371–7379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.M.; Liu, K.; Liu, J.H.; Jiang, X.L.; Wang, X.L.; Chen, Y.Z.; Li, S.G.; Zou, H.; Pang, L.J.; Liu, C.X.; et al. CD163 as a marker of M2 macrophage, contribute to predict aggressiveness and prognosis of Kazakh esophageal squamous cell carcinoma. Oncotarget 2017, 8, 21526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veloso, P.; Fernández, A.; Terraza-Aguirre, C.; Álvarez, C.; Vernal, R.; Escobar, A.; Hernández, M. Macrophages skew towards M1 profile through reduced CD163 expression in symptomatic apical periodontitis. Clin. Oral Investig. 2020, 24, 4571–4581. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Vazquez, P.A.; Bernal, L.; Paige, C.A.; Grosick, R.L.; Vilrriales, C.M.; Ferreira, D.W.; Ulecia-Morón, C.; Romero-Sandoval, E.A. Macrophage-specific nanotechnology-driven CD163 overexpression in human macrophages results in an M2 phenotype under inflammatory conditions. Immunobiology 2017, 222, 900–912. [Google Scholar] [CrossRef]
- Wang, L.; Hu, S.; Liu, Q.; Li, Y.; Xu, L.; Zhang, Z.; Cai, X.; He, X. Porcine alveolar macrophage polarization is involved in inhibition of porcine reproductive and respiratory syndrome virus (PRRSV) replication. J. Vet. Med. Sci. 2017, 79, 1906–1915. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Shi, H.; Liu, F. CD163+ M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T-cells in osteosarcoma. Int. Immunopharmacol. 2016, 34, 101–106. [Google Scholar] [CrossRef]
- Muraille, E.; Leo, O.; Moser, M. TH1/TH2 paradigm extended: Macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front. Immunol. 2014, 5, 603. [Google Scholar] [CrossRef] [Green Version]
- Shannon, A.; Carty, G.A.K. T-Cell Immunity. In Hematology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 221–239. [Google Scholar] [CrossRef]
- Nygard, A.B.; Jørgensen, C.B.; Cirera, S.; Fredholm, M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 2007, 8, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. “Trimmomatic v0.36”: Read quality filtering & adapter trimming. Bioinformatics 2014, 30, 2114–2120. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Riera, E.; Nogal, M.; Gallardo, C.; Fernández, P.; Bello-Morales, R.; López-Guerrero, J.A.; Chitko-McKown, C.G.; Richt, J.A.; Revilla, Y. Phenotyping and susceptibility of established porcine cells lines to African Swine Fever Virus infection and viral production. Sci. Rep. 2017, 7, 10369. [Google Scholar] [CrossRef]
- Rodríguez-Gómez, I.M.; Sánchez-Carvajal, J.M.; Pallarés, F.J.; Mateu, E.; Carrasco, L.; Gómez-Laguna, J. Virulent Lena strain induced an earlier and stronger downregulation of CD163 in bronchoalveolar lavage cells. Vet. Microbiol. 2019, 235, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Van Gucht, S.; Labarque, G.; Van Reeth, K. The combination of PRRS virus and bacterial endotoxin as a model for multifactorial respiratory disease in pigs. Vet. Immunol. Immunopathol. 2004, 102, 165–178. [Google Scholar] [CrossRef]
- Novakovic, P.; Harding, J.C.; Ladinig, A.; Al-Dissi, A.N.; MacPhee, D.J.; Detmer, S.E. Relationships of CD163 and CD169 positive cell numbers in the endometrium and fetal placenta with type 2 PRRSV RNA concentration in fetal thymus. Vet. Res. 2016, 47, 76. [Google Scholar] [CrossRef]
- Secombes, C.J.; Wang, T. The innate and adaptive immune system of fish. In Infectious Disease in Aquaculture; Woodhead Publishing: Shaxton, UK, 2012; pp. 3–68. [Google Scholar]
- Schmidt, A.; Oberle, N.; Krammer, P.H. Molecular mechanisms of Treg-mediated T-cell suppression. Front. Immunol. 2012, 3, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.M. Development of an Influenza Virus Vaccine Using the Baculovirus-Insect Cell Expression System: Implications for Pandemic Preparedness. 2009. Available online: https://edepot.wur.nl/14323 (accessed on 7 May 2021).
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [Green Version]
- Rayees, S.; Rochford, I.; Joshi, J.C.; Joshi, B.; Banerjee, S.; Mehta, D. Macrophage TLR4 and PAR2 signaling: Role in regulating vascular inflammatory injury and repair. Front. Immunol. 2020, 11, 2091. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Seo, H.W.; Park, C.; Chae, C. Vaccination of sows against type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) before artificial insemination protects against type 2 PRRSV challenge but does not protect against type 1 PRRSV challenge in late gestation. Vet. Res. 2014, 45, 12. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Trible, B.R.; Kerrigan, M.A.; Tian, K.; Rowland, R.R. ORF5 of porcine reproductive and respiratory syndrome virus (PRRSV) is a target of diversifying selection as infection progresses from acute infection to virus rebound. Infect. Genet. Evol. 2016, 40, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Sewell, H.F.; Agius, R.M.; Kendrick, D.; Stewart, M. COVID-19 Vaccines: Delivering Protective Immunity. BMJ 2020, 371, m4838. [Google Scholar] [CrossRef] [PubMed]
- Nazimek, K.; Ptak, W.; Nowak, B.; Ptak, M.; Askenase, P.W.; Bryniarski, K. Macrophages play an essential role in antigen-specific immune suppression mediated by T CD8+ cell-derived exosomes. Immunology 2015, 146, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Mills, C. M1 and M2 macrophages: Oracles of health and disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [Green Version]
- Mills, C.D.; Lenz, L.L.; Harris, R. A breakthrough: Macrophages-directed cancer immunotherapy. Cancer Res. 2016, 76, 513–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y. M1 and M2 polarization of macrophages: A mini-review. Med. Biol. Sci. Eng. 2019, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Xu, X.H.; Jin, L. Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, H.; Zhang, X.; He, S.; Dong, W.; Wang, X.; Chen, Y.; Liu, X.; Guo, C. Lipopolysaccharide downregulates CD163 expression to inhibit PRRSV infection via TLR4-NF-κB pathway. Front. Microbiol. 2020, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-κB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Aramburu, J.; López-Rodríguez, C. Regulation of inflammatory functions of macrophages and T lymphocytes by NFAT5. Front. Immunol. 2019, 10, 535. [Google Scholar] [CrossRef] [PubMed]
- Buxadé, M.; Lunazzi, G.; Minguillón, J.; Iborra, S.; Berga-Bolaños, R.; Del Val, M.; Aramburu, J.; López-Rodríguez, C. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 2012, 209, 379–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tellechea, M.; Buxadé, M.; Tejedor, S.; Aramburu, J.; López-Rodríguez, C. NFAT5-regulated macrophage polarization supports the proinflammatory function of macrophages and T lymphocytes. J. Immunol. 2018, 200, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Conley, J.M.; Gallagher, M.P.; Berg, L.J. T-cells and gene regulation: The switching on and turning up of genes after T-cell receptor stimulation in CD8 T-cells. Front. Immunol. 2016, 7, 76. [Google Scholar] [CrossRef] [Green Version]
- Park, S.G.; Hwang, J.R.; Byeon, Y.; Kim, D. Recent insights of T-cell receptor-mediated signaling pathways for T-cell activation and development. Exp. Mol. Med. 2020, 52, 750–761. [Google Scholar]
- Katagiri, K.; Hattori, M.; Minato, N.; Kinashi, T. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell. Biol. 2002, 22, 1001–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, L.; Emmerson, E.; Williams, H.; Saville, C.R.; Krust, A.; Chambon, P.; Mace, K.A.; Hardman, M.J. Estrogen receptor-alpha promotes alternative macrophage activation during cutaneous repair. J. Investig. Dermatol. 2014, 134, 2447–2457. [Google Scholar] [CrossRef] [Green Version]
- Noris, M.; Remuzzi, G. Overview of complement activation and regulation. In Seminars in Nephrology; WB Saunders: Milan, Italy, 2013; Volume 33, pp. 479–492. [Google Scholar]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- Furiati, S.C.; Catarino, J.S.; Silva, M.V.; Silva, R.F.; Estevam, R.B.; Teodoro, R.B.; Pereira, S.L.; Ataide, M.; Rodrigues, V.; Rodrigues, D.B. Th1, Th17, and treg responses are differently modulated by TNF-α inhibitors and methotrexate in psoriasis patients. Sci. Rep. 2019, 9, 7526. [Google Scholar] [CrossRef] [Green Version]
- Skapenko, A.; Leipe, J.; Lipsky, P.E.; Schulze-Koops, H. The role of the T-cell in autoimmune inflammation. Arthritis Res. Ther. 2005, 7, S4–S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parida, R. Cell-Mediated Immunity in Porcine Reproductive and Respiratory Syndrome Virus. 2012. Available online: https://digitalcommons.unl.edu/vetscidiss (accessed on 27 May 2021).
- Gujer, C.; Sandgren, K.J.; Douagi, I.; Adams, W.C.; Sundling, C.; Smed-Sörensen, A.; Seder, R.A.; Hedestam, G.B.; Loré, K. IFN-α roduced by human plasmacytoid dendritic cells enhances T-cell-dependent naïve B cell differentiation. J. Leukoc. Biol. 2011, 89, 811–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friederichs, K.; Schmitz, J.; Weissenbach, M.; Heinrich, P.C.; Schaper, F. Interleukin-6-induced proliferation of pre-B cells mediated by receptor complexes lacking the SHP2/SOCS3 recruitment sites revisited. Eur. J. Biochem. 2001, 268, 6401–6407. [Google Scholar] [CrossRef]
- Nakanishi, K. Unique action of interleukin-18 on T-cells and other immune cells. Front. Immunol. 2018, 9, 763. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.W.; Omori, S.A.; Labuda, T.; Karin, M.; Rickert, R.C. IKKβ is required for peripheral B cell survival and proliferation. J. Immunol. 2003, 170, 4630–4637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burks, J.; Fleury, A.; Livingston, S.; Smith, J.P. ISG15 pathway knockdown reverses pancreatic cancer cell transformation and decreases murine pancreatic tumor growth via downregulation of PDL-1 expression. Cancer Immunol. Immunother. 2019, 68, 2029–2039. [Google Scholar] [CrossRef]
- Wood, L.; Mavinkurve, V.; Muthukumaran, G.; Malo, D.; Paterson, Y. USP18 as a novel regulator of PD-1 and IFNAR1-mediated immune dysfunction. J. Immunol. 2015, 194, 629. [Google Scholar]
- Gilbert, S.C. T-cell-inducing vaccines—What’s the future. Immunology 2012, 135, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Voß, F.; van Beek, L.F.; Schwudke, D.; Ederveen, T.H.; van Opzeeland, F.J.; Thalheim, D.; Werner, S.; de Jonge, M.I.; Hammerschmidt, S. Lipidation of Pneumococcal Antigens Leads to Improved Immunogenicity and Protection. Vaccines 2020, 8, 310. [Google Scholar] [CrossRef]
- Han, Y.W.; Kim, S.B.; Rahman, M.; Uyangaa, E.; Lee, B.M.; Kim, J.H.; Park, K.I.; Hong, J.T.; Han, S.B.; Eo, S.K. Systemic and mucosal immunity induced by attenuated Salmonella enterica serovar Typhimurium expressing ORF7 of porcine reproductive and respiratory syndrome virus. Comp. Immunol. Microbiol. Infect. Dis. 2011, 34, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, M.P. PRRS immunology: What are we missing. Proc. AASV 2004, 359, 367. [Google Scholar]
- Kimman, T.G.; Cornelissen, L.A.; Moormann, R.J.; Rebel, J.M.; Stockhofe-Zurwieden, N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 2009, 27, 3704–3718. [Google Scholar] [CrossRef]
- Rompato, G.; Ling, E.; Chen, Z.; Van Kruiningen, H.; Garmendia, A.E. Positive inductive effect of IL-2 on virus-specific cellular responses elicited by a PRRSV-ORF7 DNA vaccine in swine. Vet. Immunol. Immunopathol. 2006, 109, 151–160. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequences (5′–3′) |
|---|---|
| Porcine IL-6 | GCTGCTTCTGGTGATGGCTACTGCC |
| TGAAACTCCACAAGACCGGTGGTGA | |
| Porcine TNF-α | ATGAGCACTGAGAGCATGATCCG |
| CCTCGAAGTGCAGTAGGCAGA | |
| Porcine Arg-1 | AGCCCGTGTCAACATGACTTCC |
| TTGTGTTGGCATCTTTACTGA | |
| Porcine IL-12 | CTCCCACACCGAAGCTTGAA |
| TTCTTCACCATGGGGGCT | |
| Porcine β-actin | ACAGACAGCCGTGTGTTCC |
| ACCTTCACCATCGTGTCTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahyuningtyas, R.; Lai, Y.-S.; Wu, M.-L.; Chen, H.-W.; Chung, W.-B.; Chaung, H.-C.; Chang, K.-T. Recombinant Antigen of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-2) Promotes M1 Repolarization of Porcine Alveolar Macrophages and Th1 Type Response. Vaccines 2021, 9, 1009. https://doi.org/10.3390/vaccines9091009
Wahyuningtyas R, Lai Y-S, Wu M-L, Chen H-W, Chung W-B, Chaung H-C, Chang K-T. Recombinant Antigen of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-2) Promotes M1 Repolarization of Porcine Alveolar Macrophages and Th1 Type Response. Vaccines. 2021; 9(9):1009. https://doi.org/10.3390/vaccines9091009
Chicago/Turabian StyleWahyuningtyas, Rika, Yin-Siew Lai, Mei-Li Wu, Hsin-Wei Chen, Wen-Bin Chung, Hso-Chi Chaung, and Ko-Tung Chang. 2021. "Recombinant Antigen of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-2) Promotes M1 Repolarization of Porcine Alveolar Macrophages and Th1 Type Response" Vaccines 9, no. 9: 1009. https://doi.org/10.3390/vaccines9091009
APA StyleWahyuningtyas, R., Lai, Y.-S., Wu, M.-L., Chen, H.-W., Chung, W.-B., Chaung, H.-C., & Chang, K.-T. (2021). Recombinant Antigen of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-2) Promotes M1 Repolarization of Porcine Alveolar Macrophages and Th1 Type Response. Vaccines, 9(9), 1009. https://doi.org/10.3390/vaccines9091009






