Plant-Expressed Receptor Binding Domain of the SARS-CoV-2 Spike Protein Elicits Humoral Immunity in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Agroinfiltration of N. benthamiana
2.3. Protein Expression Analysis
2.4. Purification of Plant-Based RBD
2.5. Protein Deglycosylation
2.6. LTQ-Orbitrap Mass Spectrometry
2.7. Mice and Immunization Protocol
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. Focus Reduction Neutralization Test (FRNT)
2.10. Statistical Analysis
3. Results
3.1. Transient Expression of RBD in N. benthamiana
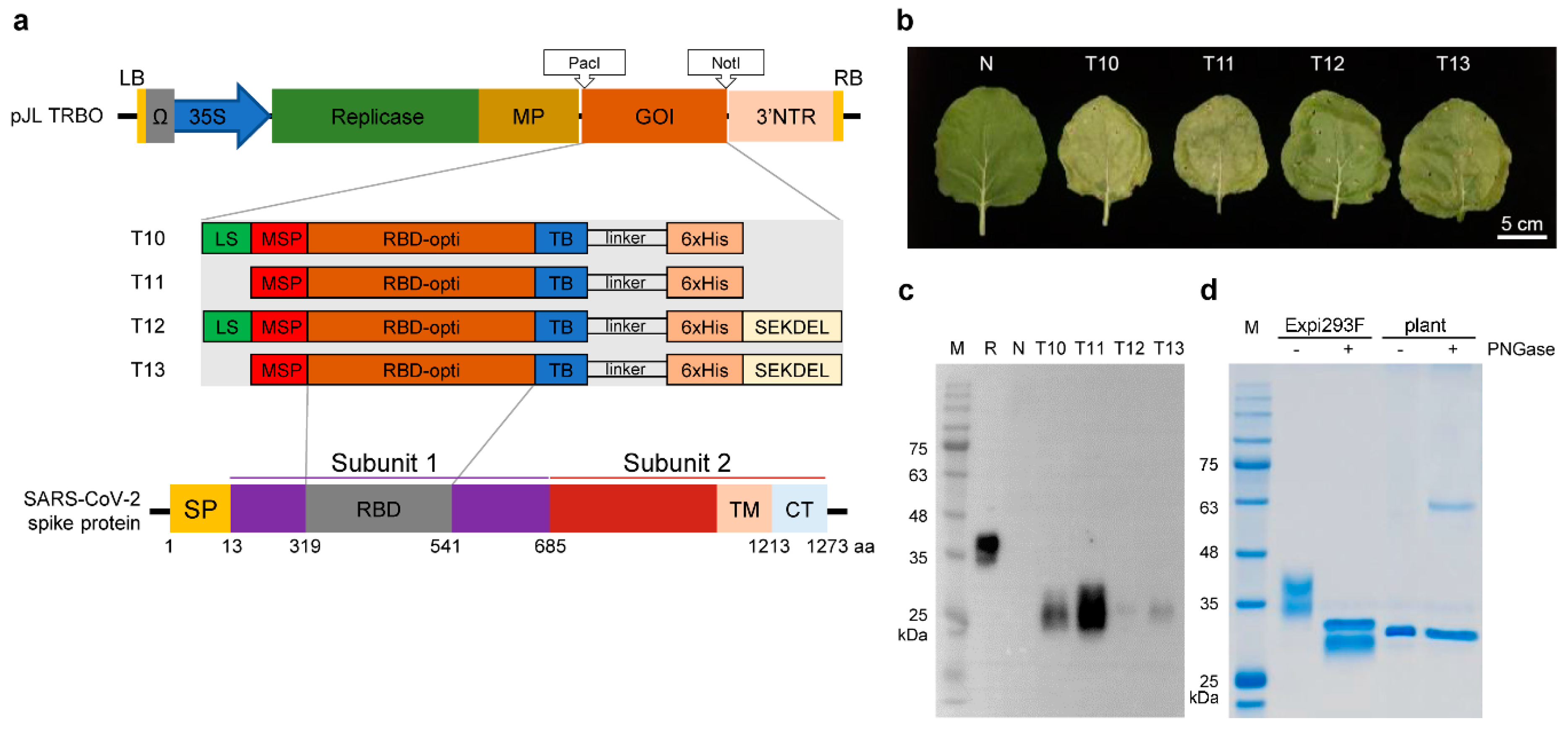
3.2. Purification of Plant-Expressed RBD
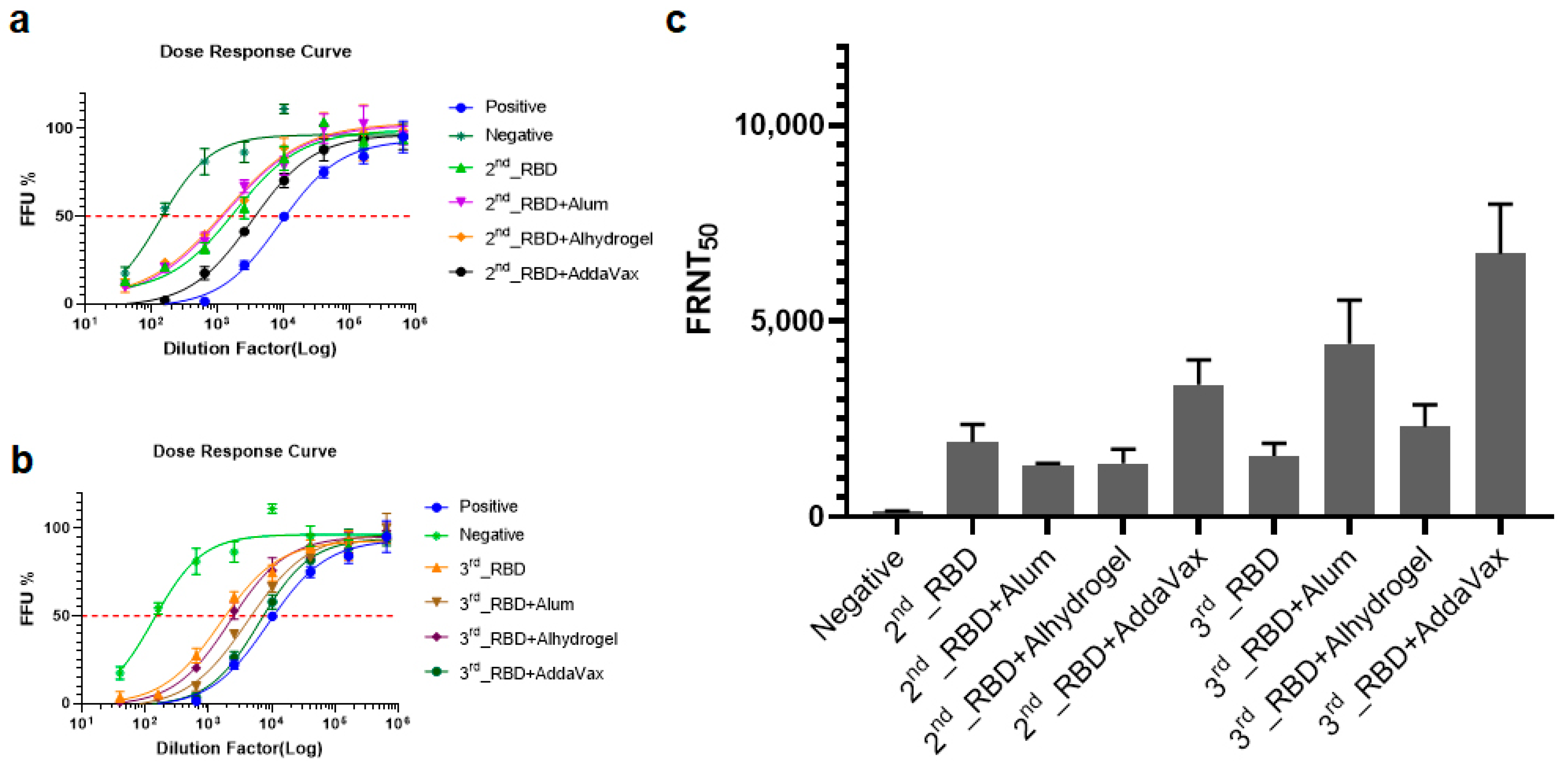
3.3. Plant-Based RBD Induces a Humoral Immune Response in Mice and Protects Vero E6 Cells from SARS-CoV-2 Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus Disease (COVID-19). Available online: https://covid19.who.int/table (accessed on 12 June 2021).
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- World Health Organization. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS); World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- WHO. Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Available online: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1 (accessed on 12 June 2021).
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.H.; Fung, K.; Choy, K.-T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- D’Aoust, M.-A.; Couture, M.M.-J.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.-P. The production of hemagglutinin-based virus-like particles in plants: A rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 2010, 8, 607–619. [Google Scholar] [CrossRef]
- Sil, B.; Jha, S. Plants: The Future Pharmaceutical Factory. Am. J. Plant Sci. 2014, 5, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Shim, B.-S.; Hong, K.-J.; Maharjan, P.M.; Choe, S. Plant factory: New resource for the productivity and diversity of human and veterinary vaccines. Clin. Exp. Vaccine Res. 2019, 8, 136–139. [Google Scholar] [CrossRef]
- Thanavala, Y.; Mahoney, M.; Pal, S.; Scott, A.; Richter, L.; Natarajan, N.; Goodwin, P.; Arntzen, C.J.; Mason, H.S. Immunogenicity in humans of an edible vaccine for hepatitis B. Proc. Natl. Acad. Sci. USA 2005, 102, 3378–3382. [Google Scholar] [CrossRef] [Green Version]
- Bhairy, S.R.; Hirlekar, R. Edible vaccines: An advancement in oral immunization. Asian J. Pharm. Clin. Res. 2017, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Hiatt, A.; Caffferkey, R.; Bowdish, K. Production of antibodies in transgenic plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef]
- Marillonnet, S.; Thoeringer, C.; Kandzia, R.; Klimyuk, V.; Gleba, Y. Systemic Agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005, 23, 718–723. [Google Scholar] [CrossRef]
- Shaaltiel, Y.; Bartfeld, D.; Hashmueli, S.; Baum, G.; Brill-Almon, E.; Galili, G.; Dym, O.; Boldin-Adamsky, S.A.; Silman, I.; Sussman, J.; et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher’s disease using a plant cell system. Plant Biotechnol. J. 2007, 5, 579–590. [Google Scholar] [CrossRef]
- Montero-Morales, L.; Steinkellner, H. Advanced Plant-Based Glycan Engineering. Front. Bioeng. Biotechnol. 2018, 6, 81. [Google Scholar] [CrossRef]
- Diamos, A.G.; Rosenthal, S.H.; Mason, H.S. 5′ and 3′ Untranslated Regions Strongly Enhance Performance of Geminiviral Replicons in Nicotiana benthamiana Leaves. Front. Plant Sci. 2016, 7, 200. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Weng, Y.; Dickey, A.; Wang, K.Y. Plants as Factories for Human Pharmaceuticals: Applications and Challenges. Int. J. Mol. Sci. 2015, 16, 28549–28565. [Google Scholar] [CrossRef]
- Hefferon, K.L. DNA Virus Vectors for Vaccine Production in Plants: Spotlight on Geminiviruses. Vaccines 2014, 2, 642. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Ma, J.K.-C. Plant-made pharmaceuticals: Leading products and production platforms. Biotechnol. Appl. Biochem. 2011, 58, 58–67. [Google Scholar] [CrossRef]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical Analysis of the Commercial Potential of Plants for the Production of Recombinant Proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, P.M.; Choe, S. Transient expression of hemagglutinin antigen from canine influenza virus H3N2 in Nicotiana benthamiana and Lactuca sativa. Clin. Exp. Vaccine Res. 2019, 8, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Buyel, J. Molecular farming—The slope of enlightenment. Biotechnol. Adv. 2020, 40, 107519. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Rattanapisit, K.; Manopwisedjaroen, S.; Thitithanyanont, A.; Phoolcharoen, W. Monoclonal Antibodies B38 and H4 Produced in Nicotiana benthamiana Neutralize SARS-CoV-2 in vitro. Front. Plant Sci. 2020, 11, 589995. [Google Scholar] [CrossRef]
- Donini, M.; Marusic, C. Current state-of-the-art in plant-based antibody production systems. Biotechnol. Lett. 2019, 41, 335–346. [Google Scholar] [CrossRef]
- Mor, T.S. Molecular pharming’s foot in the FDA’s door: Protalix’s trailblazing story. Biotechnol. Lett. 2015, 37, 2147–2150. [Google Scholar] [CrossRef] [Green Version]
- Takeyama, N.; Kiyono, H.; Yuki, Y. Plant-based vaccines for animals and humans: Recent advances in technology and clinical trials. Ther. Adv. Vaccines 2015, 3, 139–154. [Google Scholar] [CrossRef]
- Mulangu, S.; Dodd, L.E.; Davey, R.T.; Mbaya, O.T.; Proschan, M.; Mukadi, D.; Manzo, M.L.; Nzolo, D.; Oloma, A.T.; Ibanda, A.; et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381, 2293–2303. [Google Scholar] [CrossRef]
- Clinicaltrials. Investigational Therapeutics for the Treatment of People with Ebola Virus Disease. Available online: https://clinicaltrials.gov/ct2/show/NCT03719586 (accessed on 12 June 2021).
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): Two multicentre, randomised phase 3 trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Medicago. Medicago Pipeline. Available online: https://www.medicago.com/en/pipeline (accessed on 12 June 2021).
- Capell, T.; Twyman, R.M.; Armario-Najera, V.; Ma, J.K.-C.; Schillberg, S.; Christou, P. Potential Applications of Plant Biotechnology against SARS-CoV-2. Trends Plant Sci. 2020, 25, 635–643. [Google Scholar] [CrossRef]
- McDonald, K.A.; Holtz, R.B. From Farm to Finger Prick—A Perspective on How Plants Can Help in the Fight Against COVID-19. Front. Bioeng. Biotechnol. 2020, 8, 782. [Google Scholar] [CrossRef]
- Dhama, K.; Natesan, S.; Yatoo, M.I.; Patel, S.K.; Tiwari, R.; Saxena, S.K.; Harapan, H. Plant-based vaccines and antibodies to combat COVID-19: Current status and prospects. Hum. Vaccines Immunother. 2020, 16, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Márquez-Escobar, V.A.; González-Ortega, O.; Nieto-Gómez, R.; Arévalo-Villalobos, J.I. What Does Plant-Based Vaccine Technology Offer to the Fight against COVID-19? Vaccines 2020, 8, 183. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, Z.; Waterhouse, P.; Bally, J. Plant-Based Vaccines: The Way Ahead? Viruses 2020, 13, 5. [Google Scholar] [CrossRef]
- Phoolcharoen, W.; Shanmugaraj, B. Addressing demand for recombinant biopharmaceuticals in the COVID-19 era. Asian Pac. J. Trop. Med. 2021, 14, 49. [Google Scholar] [CrossRef]
- Kumar, A.U.; Kadiresen, K.; Gan, W.C.; Ling, A.P.K. Current updates and research on plant-based vaccines for coronavirus disease. Clin. Exp. Vaccine Res. 2021, 10, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Rattanapisit, K.; Yusakul, G.; Shanmugaraj, B.; Kittirotruji, K.; Suwatsrisakul, P.; Prompetchara, E.; Taychakhoonavud, S.; Phoolcharoen, W. Plant-produced recombinant SARS-CoV-2 receptorbinding domain; an economical, scalable biomaterial source for COVID-19 diagnosis. Biomater. Transl. 2021, 2, 43–49. [Google Scholar]
- Clinicaltrials. Study of a Recombinant Coronavirus-like Particle COVID-19 Vaccine in Adults. Available online: https://clinicaltrials.gov/ct2/show/NCT04636697?term=Medicago+COVID-19&draw=2&rank=1 (accessed on 12 June 2021).
- Medicago. Medicago and GSK Start Phase 3 Trial of Adjuvanted COVID-19 Vaccine Candidate. Available online: https://www.medicago.com/en/media-room/medicago-and-gsk-start-phase-3-trial-of-adjuvanted-covid-19-vaccine-candidate/ (accessed on 12 June 2021).
- KBP. KBP-201 COVID-19 Vaccine Trial in Healthy Volunteers. Available online: https://clinicaltrials.gov/ct2/show/NCT04473690?term=KBP201&cond=Covid19&draw=2&rank=1 (accessed on 12 June 2021).
- Wolfert, M.; Boons, G.-J. Adaptive immune activation: Glycosylation does matter. Nat. Chem. Biol. 2013, 9, 776–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdilek, A.; Paschall, A.V.; Dookwah, M.; Tiemeyer, M.; Avci, F.Y. Host protein glycosylation in nucleic acid vaccines as a potential hurdle in vaccine design for nonviral pathogens. Proc. Natl. Acad. Sci. USA 2020, 117, 1280–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wintjens, R.; Bifani, A.M.; Bifani, P. Impact of glycan cloud on the B-cell epitope prediction of SARS-CoV-2 Spike protein. npj Vaccines 2020, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Castilho, A.; Stadlmann, J.; Kunert, R.; Quendler, H.; Gattinger, P.; Jez, J.; Rademacher, T.; Altmann, F.; Mach, L.; et al. Improved Virus Neutralization by Plant-produced Anti-HIV Antibodies with a Homogeneous β1,4-Galactosylated N-Glycan Profile. J. Biol. Chem. 2009, 284, 20479–20485. [Google Scholar] [CrossRef] [Green Version]
- Lindbo, J.A. TRBO: A High-Efficiency Tobacco Mosaic Virus RNA-Based Overexpression Vector. Plant Physiol. 2007, 145, 1232–1240. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-M.; Chung, Y.-S.; Jo, H.J.; Lee, N.-J.; Kim, M.S.; Woo, S.H.; Park, S.; Kim, J.W.; Kim, H.M.; Han, M.-G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020, 11, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 255. [Google Scholar] [CrossRef]
- Tee, K.L.; Philip, J.J.; Joseph, M.S.; Stephen, R.P.J.; Abayomi, O.J.; Yusuf, J.; Thilo, H.P.; Theo, M.; Joseph, P.; James, G.; et al. Purification of recombinant SARS-CoV-2 spike, its receptor binding domain, and CR3022 mAb for serological assay. bioRxiv 2020. [Google Scholar] [CrossRef]
- Di Pasquale, A.; Preiss, S.; Silva, F.M.D.O.E.; Garçon, N. Vaccine Adjuvants: From 1920 to 2015 and Beyond. Vaccines 2015, 3, 320–343. [Google Scholar] [CrossRef] [Green Version]
- Stevens, T.L.; Bossie, A.; Sanders, V.M.; Fernandez-Botran, R.; Coffman, R.L.; Mosmann, T.R.; Vitetta, E.S. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 1988, 334, 255–258. [Google Scholar] [CrossRef]
- Firacative, C.; Gressler, A.E.; Schubert, K.; Schulze, B.; Müller, U.; Brombacher, F.; Von Bergen, M.; Alber, G. Identification of T helper (Th)1- and Th2-associated antigens of Cryptococcus neoformans in a murine model of pulmonary infection. Sci. Rep. 2018, 8, 268. [Google Scholar] [CrossRef]
- Zhao, F.; Qu, J.; Wang, W.; Li, S.; Xu, S. The imbalance of Th1/Th2 triggers an inflammatory response in chicken spleens after ammonia exposure. Poult. Sci. 2020, 99, 3817–3822. [Google Scholar] [CrossRef]
- Wouters, O.J.; Shadlen, K.C.; Salcher-Konrad, M.; Pollard, A.J.; Larson, H.J.; Teerawattananon, Y.; Jit, M. Challenges in ensuring global access to COVID-19 vaccines: Production, affordability, allocation, and deployment. Lancet 2021, 397, 1023–1034. [Google Scholar] [CrossRef]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2020, 21, 73–82. [Google Scholar] [CrossRef]
- Li, T.; Zhang, Y.; Fu, L.; Yu, C.; Li, X.; Li, Y.; Zhang, X.; Rong, Z.; Wang, Y.; Ning, H.; et al. siRNA targeting the Leader sequence of SARS-CoV inhibits virus replication. Gene Ther. 2005, 12, 751–761. [Google Scholar] [CrossRef] [Green Version]
- Baldassarre, A.; Paolini, A.; Bruno, S.P.; Felli, C.; Tozzi, A.E.; Masotti, A. Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5′UTR of SARS-CoV-2. Epigenomics 2020, 12, 1349–1361. [Google Scholar] [CrossRef]
- Rattanapisit, K.; Shanmugaraj, B.; Manopwisedjaroen, S.; Purwono, P.B.; Siriwattananon, K.; Khorattanakulchai, N.; Hanittinan, O.; Boonyayothin, W.; Thitithanyanont, A.; Smith, D.R.; et al. Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 2020, 10, 17698. [Google Scholar] [CrossRef]
- Laere, E.; Ling, A.P.K.; Wong, Y.P.; Koh, R.Y.; Lila, M.A.M.; Hussein, S. Plant-Based Vaccines: Production and Challenges. J. Bot. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sabalza, M.; Christou, P.; Capell, T. Recombinant plant-derived pharmaceutical proteins: Current technical and economic bottlenecks. Biotechnol. Lett. 2014, 36, 2367–2379. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef]
- Forno, G.; Bollati-Fogolín, M.; Oggero, M.; Kratje, R.; Etcheverrigaray, M.; Conradt, H.S.; Nimtz, M. N- and O-linked carbohydrates and glycosylation site occupancy in recombinant human granulocyte-macrophage colony-stimulating factor secreted by a Chinese hamster ovary cell line. JBIC J. Biol. Inorg. Chem. 2004, 271, 907–919. [Google Scholar] [CrossRef]
- Makatsa, M.S.; Tincho, M.B.; Wendoh, J.M.; Ismail, S.D.; Nesamari, R.; Pera, F.; de Beer, S.; David, A.; Jugwanth, S.; Gededzha, M.P.; et al. SARS-CoV-2 Antigens Expressed in Plants Detect Antibody Responses in COVID-19 Patients. Front. Plant Sci. 2021, 12, 589940. [Google Scholar] [CrossRef]
- Kiros, T.G.; Levast, B.; Auray, G.; Strom, S.; Van Kessel, J.; Gerdts, V. The Importance of Animal Models in the Development of Vaccines. In Innovation in Vaccinology: From Design, Through to Delivery and Testing; Baschieri, S., Ed.; Springer: Heidelberg, Germany, 2012; pp. 251–264. [Google Scholar] [CrossRef]
- Justice, M.J.; Dhillon, P. Using the mouse to model human disease: Increasing validity and reproducibility. Dis. Model. Mech. 2016, 9, 101–103. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H. BALB/c Mouse. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 290–292. [Google Scholar]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Hütter, J.; Rödig, J.V.; Höper, D.; Seeberger, P.H.; Reichl, U.; Rapp, E.; Lepenies, B. Toward Animal Cell Culture–Based Influenza Vaccine Design: Viral HemagglutininN-Glycosylation Markedly Impacts Immunogenicity. J. Immunol. 2012, 190, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowling, W.; Thompson, E.; Badger, C.; Mellquist, J.L.; Garrison, A.R.; Smith, J.M.; Paragas, J.; Hogan, R.J.; Schmaljohn, C. Influences of Glycosylation on Antigenicity, Immunogenicity, and Protective Efficacy of Ebola Virus GP DNA Vaccines. J. Virol. 2007, 81, 1821–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitter, J.N.; Means, R.E.; Desrosiers, R.C. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 1998, 4, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Sugimoto, C.; Ohgimoto, S.; Nakayama, E.E.; Shioda, T.; Kusagawa, S.; Takebe, Y.; Kano, M.; Matano, T.; Yuasa, T.; et al. Influence of Glycosylation on the Efficacy of an Env-Based Vaccine against Simian Immunodeficiency Virus SIVmac239 in a Macaque AIDS Model. J. Virol. 2005, 79, 10386–10396. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-M.; Quan, F.S.; Huang, C.; Guo, L.; Ye, L.; Yang, C.; Compans, R.W. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 2004, 331, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, K.S.; Steckbeck, J.D.; Rowles, J.L.; Desrosiers, R.C.; Montelaro, R.C. Removal of N-Linked Glycosylation Sites in the V1 Region of Simian Immunodeficiency Virus gp120 Results in Redirection of B-Cell Responses to V3. J. Virol. 2004, 78, 1525–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, L.; Wei, J.; Liu, Z.; Keegan, B.; Fujiwara, R.T.; Jones, K.M.; Asojo, O.; Strych, U.; Bottazzi, M.E.; Hotez, P.J.; et al. Protective immunity elicited by the nematode-conserved As37 recombinant protein against Ascaris suum infection. PLoS Negl. Trop. Dis. 2020, 14, e0008057. [Google Scholar] [CrossRef]
- Choubini, E.; Habibi, M.; Khorshidi, A.; Ghasemi, A.; Karam, M.R.A.; Bouzari, S. A novel multi-peptide subunit vaccine admixed with AddaVax adjuvant produces significant immunogenicity and protection against Proteus mirabilis urinary tract infection in mice model. Mol. Immunol. 2018, 96, 88–97. [Google Scholar] [CrossRef]
- Du, Y.; Xu, Y.; Feng, J.; Hu, L.; Zhang, Y.; Zhang, B.; Guo, W.; Mai, R.; Chen, L.; Fang, J.; et al. Intranasal administration of a recombinant RBD vaccine induced protective immunity against SARS-CoV-2 in mouse. Vaccine 2021, 39, 2280–2287. [Google Scholar] [CrossRef]
- Baisa, G.; Rancour, D.; Mansfield, K.; Burns, M.; Martin, L.; Cunha, D.; Fischer, J.; Muecksch, F.; Hatziioannou, T.; Bieniasz, P.D.; et al. A Recombinant Protein SARS-CoV-2 Candidate Vaccine Elicits High-titer Neutralizing Antibodies in Macaques. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Sun, S.; He, L.; Zhao, Z.; Gu, H.; Fang, X.; Wang, T.; Yang, X.; Chen, S.; Deng, Y.; Li, J.; et al. Recombinant vaccine containing an RBD-Fc fusion induced protection against SARS-CoV-2 in nonhuman primates and mice. Cell. Mol. Immunol. 2021, 18, 1070–1073. [Google Scholar] [CrossRef] [PubMed]

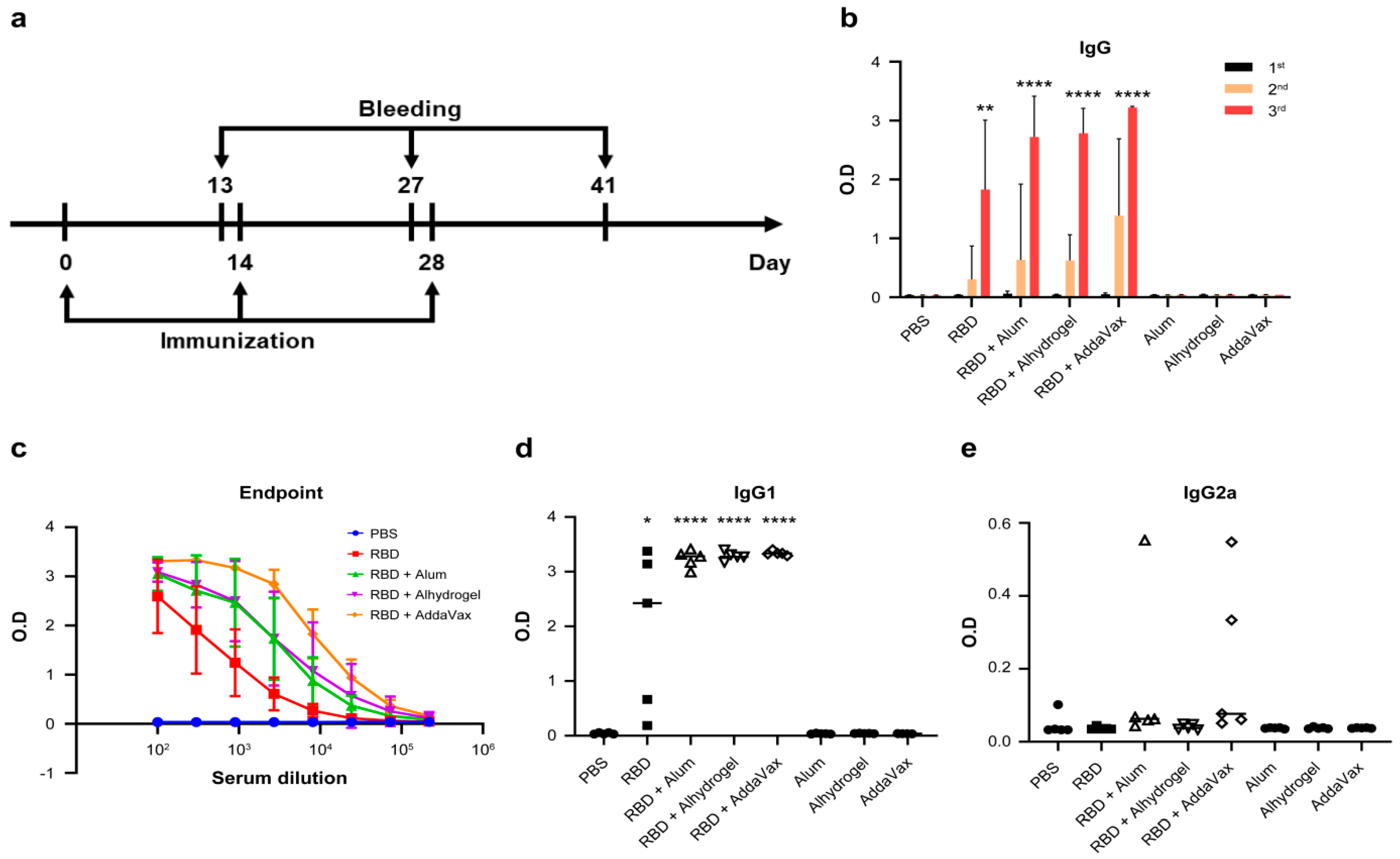
| No. | Group | Mouse No. | Antigen | Adjuvant | Total Volume | Immunization Route |
|---|---|---|---|---|---|---|
| 1 | PBS | 5 | - | - | ||
| 2 | RBD only | 5 | 10 μg of RBD | - | 50 µL | I.M. |
| 3 | RBD + Alum | 5 | 10 μg of RBD | Alum | ||
| 4 | RBD + Alhydrogel | 5 | 10 μg of RBD | Alhydrogel | ||
| 5 | RBD + AddaVax | 5 | 10 μg of RBD | AddaVax | ||
| 6 | Alum only | 5 | - | Alum | ||
| 7 | Alhydrogel only | 5 | - | Alhydrogel | ||
| 8 | AddaVax only | 5 | - | AddaVax |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maharjan, P.M.; Cheon, J.; Jung, J.; Kim, H.; Lee, J.; Song, M.; Jeong, G.U.; Kwon, Y.; Shim, B.; Choe, S. Plant-Expressed Receptor Binding Domain of the SARS-CoV-2 Spike Protein Elicits Humoral Immunity in Mice. Vaccines 2021, 9, 978. https://doi.org/10.3390/vaccines9090978
Maharjan PM, Cheon J, Jung J, Kim H, Lee J, Song M, Jeong GU, Kwon Y, Shim B, Choe S. Plant-Expressed Receptor Binding Domain of the SARS-CoV-2 Spike Protein Elicits Humoral Immunity in Mice. Vaccines. 2021; 9(9):978. https://doi.org/10.3390/vaccines9090978
Chicago/Turabian StyleMaharjan, Puna Maya, Jinyeong Cheon, Jiyun Jung, Haerim Kim, Jaewon Lee, Minjeong Song, Gi Uk Jeong, Youngchan Kwon, Byoungshik Shim, and Sunghwa Choe. 2021. "Plant-Expressed Receptor Binding Domain of the SARS-CoV-2 Spike Protein Elicits Humoral Immunity in Mice" Vaccines 9, no. 9: 978. https://doi.org/10.3390/vaccines9090978
APA StyleMaharjan, P. M., Cheon, J., Jung, J., Kim, H., Lee, J., Song, M., Jeong, G. U., Kwon, Y., Shim, B., & Choe, S. (2021). Plant-Expressed Receptor Binding Domain of the SARS-CoV-2 Spike Protein Elicits Humoral Immunity in Mice. Vaccines, 9(9), 978. https://doi.org/10.3390/vaccines9090978






