Obtaining Precise Molecular Information via DNA Nanotechnology
Abstract
:1. Introduction
2. DNA Nanotechnology
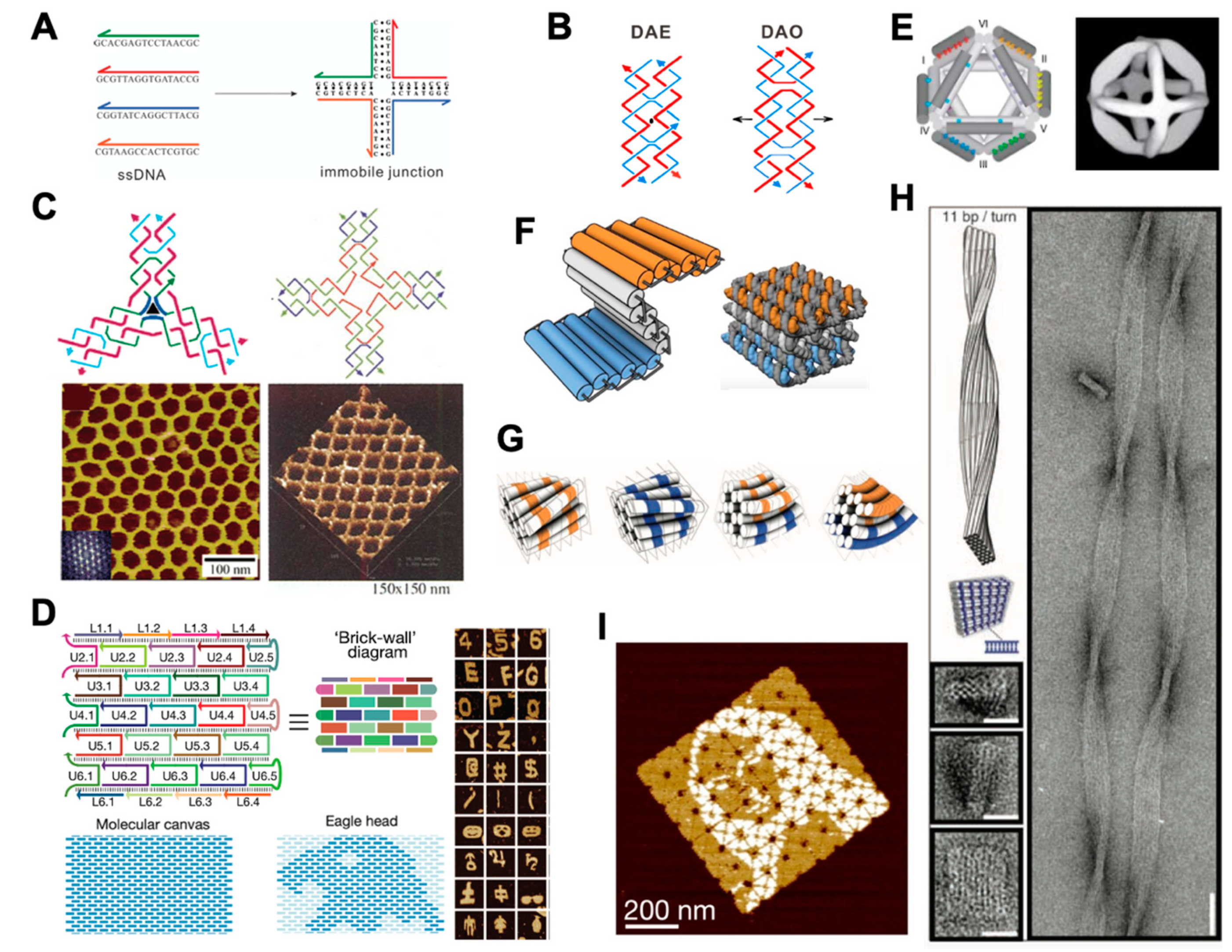
2.1. Structural DNA Nanotechnology
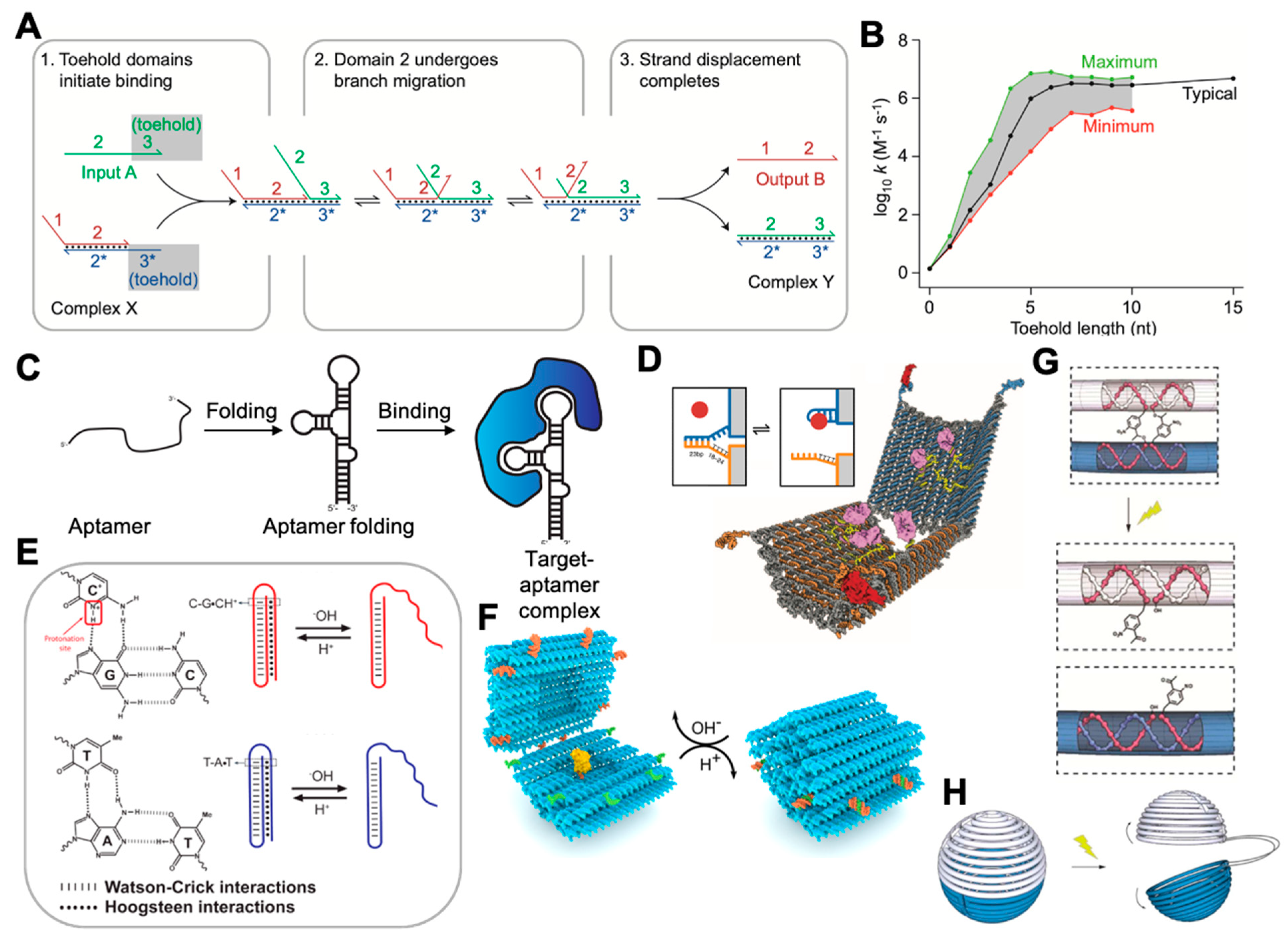
2.2. Dynamic DNA Nanotechnology
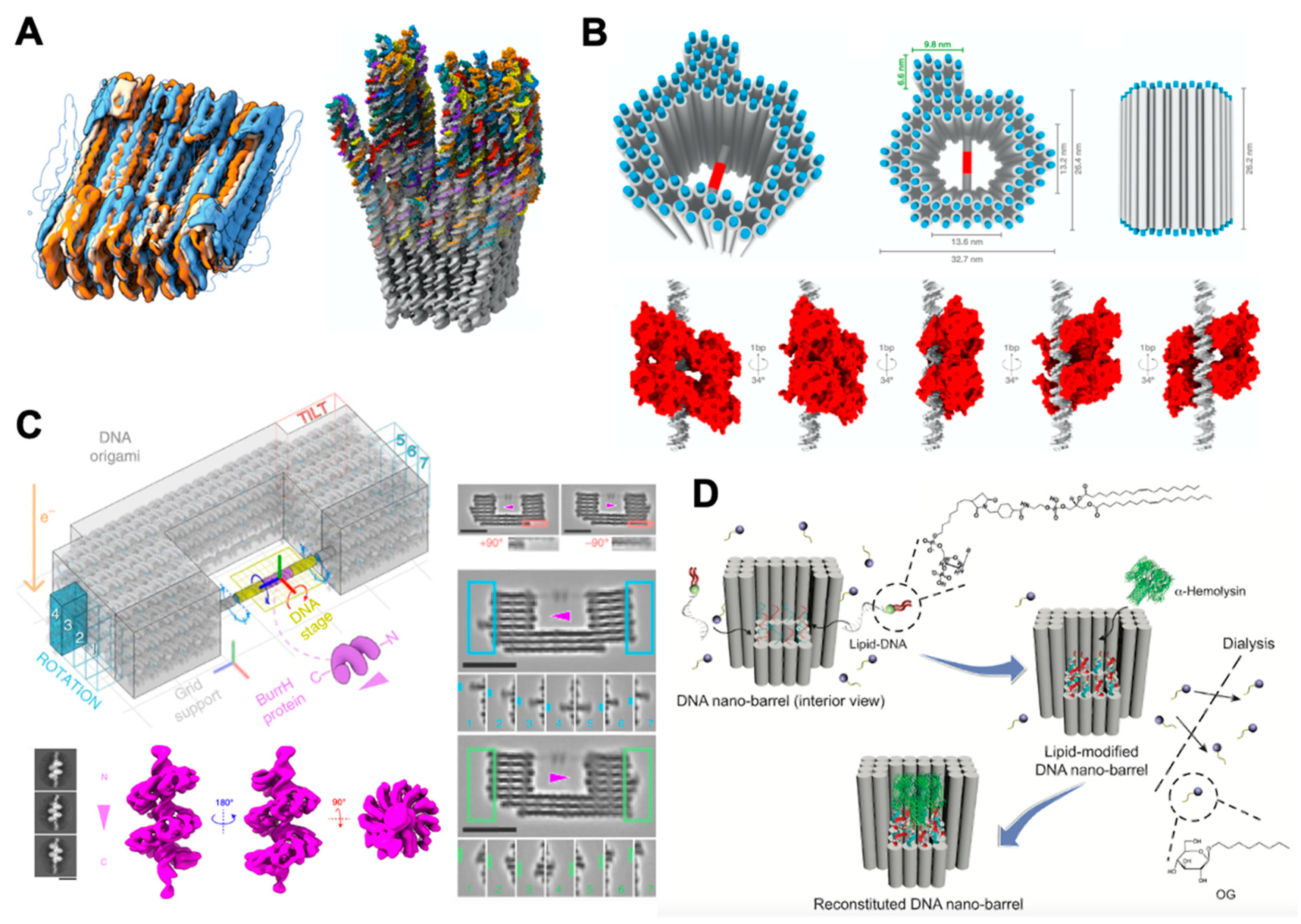
3. DNA Nanotechnology for Structural Reconstruction of Proteins
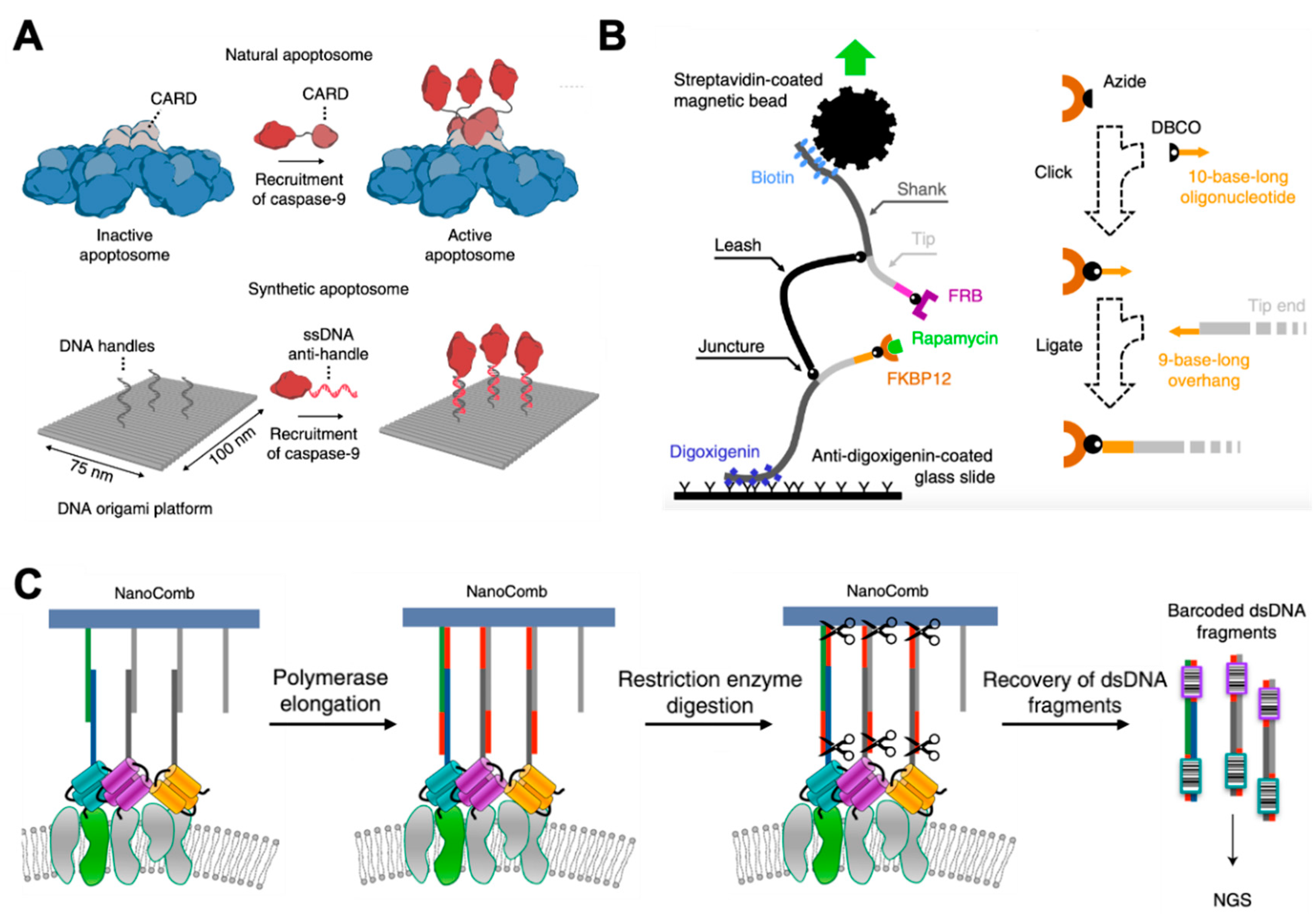
4. Study of Protein–Protein Interactions
5. Measurement of Molecular Forces
6. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, X.-C.; McMullan, G.; Scheres, S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015, 40, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y. Single-particle cryo-EM—How did it get here and where will it go. Science 2018, 361, 876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellenkamp, B.; Schmid, S.; Doroshenko, O.; Opanasyuk, O.; Kühnemuth, R.; Rezaei Adariani, S.; Ambrose, B.; Aznauryan, M.; Barth, A.; Birkedal, V.; et al. Precision and accuracy of single-molecule FRET measurements—Multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Ecken, J.v.d.; Heissler, S.M.; Pathan-Chhatbar, S.; Manstein, D.J.; Raunser, S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 2016, 534, 724–728. [Google Scholar] [CrossRef]
- Thakur, A.K.; Movileanu, L. Real-time measurement of protein–protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 2019, 37, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Asher, W.B.; Geggier, P.; Holsey, M.D.; Gilmore, G.T.; Pati, A.K.; Meszaros, J.; Terry, D.S.; Mathiasen, S.; Kaliszewski, M.J.; McCauley, M.D.; et al. Single-molecule FRET imaging of GPCR dimers in living cells. Nat. Methods 2021, 18, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Wang, Z.; Qu, L.; Huang, W.; Guo, S.; Guan, X.; Zhang, W.; Sun, F.; Yuan, H.; Zou, H.; et al. Single-molecule multiplexed profiling of protein-DNA complexes using magnetic tweezers. J. Biol. Chem. 2021, 296. [Google Scholar] [CrossRef]
- Martin, T.G.; Bharat, T.A.M.M.; Joerger, A.C.; Bai, X.-c.C.; Praetorius, F.; Fersht, A.R.; Dietz, H.; Scheres, S.H.W.W. Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. USA 2016, 113, E7456–E7463. [Google Scholar] [CrossRef] [Green Version]
- Ambrosetti, E.; Bernardinelli, G.; Hoffecker, I.; Hartmanis, L.; Kiriako, G.; de Marco, A.; Sandberg, R.; Högberg, B.; Teixeira, A.I. A DNA-nanoassembly-based approach to map membrane protein nanoenvironments. Nat. Nanotechnol. 2021, 16, 85–95. [Google Scholar] [CrossRef]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, S.; An, B.; Huang, K.; Bai, T.; Xu, M.; Bellot, G.; Ke, Y.; Xiang, Y.; Wei, B. Complex wireframe DNA nanostructures from simple building blocks. Nat. Commun. 2019, 10, 1067. [Google Scholar] [CrossRef] [Green Version]
- Praetorius, F.; Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 2017, 355, eaam5488. [Google Scholar] [CrossRef]
- Xin, L.; Duan, X.; Liu, N. Dimerization and oligomerization of DNA-assembled building blocks for controlled multi-motion in high-order architectures. Nat. Commun. 2021, 12, 3207. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Schreiber, R.; Zhang, H.; Govorov, A.O.; Liedl, T.; Liu, N. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 2014, 13, 862–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, G.; Zhang, F.; Wang, F.; Peng, T.; Liu, H.; Poppleton, E.; Šulc, P.; Jiang, S.; Liu, L.; Gong, C.; et al. Meta-DNA structures. Nat. Chem. 2020, 12, 1067–1075. [Google Scholar] [CrossRef]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Ong, L.L.; Sun, W.; Song, J.; Dong, M.; Shih, W.M.; Yin, P. DNA brick crystals with prescribed depths. Nat. Chem. 2014, 6, 994–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, S.F.J.; Auer, A.; Min, J.; Ponnuswamy, N.; Woehrstein, J.B.; Schueder, F.; Strauss, M.T.; Schnitzbauer, J.; Nathwani, B.; Zhao, Z.; et al. Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard. Nat. Commun. 2020, 11, 5768. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, F.; Jing, X.; Pan, M.; Liu, P.; Li, W.; Zhu, B.; Li, J.; Chen, H.; Wang, L.; et al. Complex silica composite nanomaterials templated with DNA origami. Nature 2018, 559, 593–598. [Google Scholar] [CrossRef]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar] [CrossRef]
- Pinheiro, A.V.; Han, D.; Shih, W.M.; Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. [Google Scholar] [CrossRef]
- Hernandez-Garcia, A. Strategies to Build Hybrid Protein–DNA Nanostructures. Nanomaterials 2021, 11, 1332. [Google Scholar] [CrossRef]
- Stephanopoulos, N.; Šulc, P. DNA Nanodevices as Mechanical Probes of Protein Structure and Function. Appl. Sci. 2021, 11, 2802. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Bednar, J.; Furrer, P.; Katritch, V.; Stasiak, A.; Dubochet, J.; Stasiak, A. Determination of DNA Persistence Length by Cryo-electron Microscopy. Separation of the Static and Dynamic Contributions to the Apparent Persistence Length of DNA. J. Mol. Biol. 1995, 254, 579–594. [Google Scholar] [CrossRef]
- Dirks, R.M.; Bois, J.S.; Schaeffer, J.M.; Winfree, E.; Pierce, N.A. Thermodynamic Analysis of Interacting Nucleic Acid Strands. SIAM Rev. 2007, 49, 65–88. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.X.; Fang, J.Z.; Duan, W.; Wu, L.R.; Zhang, A.W.; Dalchau, N.; Yordanov, B.; Petersen, R.; Phillips, A.; Zhang, D.Y. Predicting DNA hybridization kinetics from sequence. Nat. Chem. 2018, 10, 91–98. [Google Scholar] [CrossRef] [Green Version]
- SantaLucia, J.; Hicks, D. The Thermodynamics of DNA Structural Motifs. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 415–440. [Google Scholar] [CrossRef] [Green Version]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Srinivas, N.; Ouldridge, T.E.; Šulc, P.; Schaeffer, J.M.; Yurke, B.; Louis, A.A.; Doye, J.P.K.; Winfree, E. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 2013, 41, 10641–10658. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Kallenbach, N.R.; Ma, R.-I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 1983, 305, 829–831. [Google Scholar] [CrossRef]
- Fu, T.J.; Seeman, N.C. DNA double-crossover molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Liu, H.; Ribbe, A.E.; Mao, C. Self-Assembly of Hexagonal DNA Two-Dimensional (2D) Arrays. J. Am. Chem. Soc. 2005, 127, 12202–12203. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 2004, 427, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Tikhomirov, G.; Petersen, P.; Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. [Google Scholar] [CrossRef] [Green Version]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, Analysis, Ligation, and Self-Assembly of DNA Triple Crossover Complexes. J. Am. Chem. Soc. 2000, 122, 1848–1860. [Google Scholar] [CrossRef]
- Shen, Z.; Yan, H.; Wang, T.; Seeman, N.C. Paranemic Crossover DNA: A Generalized Holliday Structure with Applications in Nanotechnology. J. Am. Chem. Soc. 2004, 126, 1666–1674. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Sha, R.; Seeman, N.C. Pseudohexagonal 2D DNA Crystals from Double Crossover Cohesion. J. Am. Chem. Soc. 2004, 126, 10230–10231. [Google Scholar] [CrossRef]
- Yin, P.; Hariadi, R.F.; Sahu, S.; Choi, H.M.T.; Park, S.H.; LaBean, T.H.; Reif, J.H. Programming DNA Tube Circumferences. Science 2008, 321, 824. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, Y.; Douglas, S.M.; Liu, M.; Sharma, J.; Cheng, A.; Leung, A.; Liu, Y.; Shih, W.M.; Yan, H. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A three-dimensional editing tool for complex DNA structures. In Proceedings of the DNA Computing, Berlin/Heidelberg, Germany, 8–11 June 2009; pp. 90–101. [Google Scholar]
- Doty, D.; Lee, B.L.; Stérin, T. Scadnano: A browser-based, scriptable tool for designing DNA nanostructures. arXiv 2020, arXiv:2005.11841. [Google Scholar]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, E.; Mohammed, A.; Bosco, A.; Teixeira, A.I.; Orponen, P.; Högberg, B. Computer-Aided Production of Scaffolded DNA Nanostructures from Flat Sheet Meshes. Angew. Chem. Int. Ed. 2016, 55, 8869–8872. [Google Scholar] [CrossRef] [PubMed]
- Jun, H.; Zhang, F.; Shepherd, T.; Ratanalert, S.; Qi, X.; Yan, H.; Bathe, M. Autonomously designed free-form 2D DNA origami. Sci. Adv. 2019, 5, eaav0655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, H.; Wang, X.; Bricker, W.P.; Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 2019, 10, 5419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Winfree, E. Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J. Am. Chem. Soc. 2009, 131, 17303–17314. [Google Scholar] [CrossRef] [Green Version]
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A cargo-sorting DNA robot. Science 2017, 357. [Google Scholar] [CrossRef] [Green Version]
- Sherman, W.B.; Seeman, N.C. A Precisely Controlled DNA Biped Walking Device. Nano Lett. 2004, 4, 1203–1207. [Google Scholar] [CrossRef]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef] [Green Version]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275. [Google Scholar] [CrossRef] [Green Version]
- Seelig, G.; Soloveichik, D.; Zhang, D.Y.; Winfree, E. Enzyme-Free Nucleic Acid Logic Circuits. Science 2006, 314, 1585. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Winfree, E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332, 1196. [Google Scholar] [CrossRef] [Green Version]
- Lopez, R.; Wang, R.; Seelig, G. A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 2018, 10, 746–754. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Xu, X.; Xu, R.; Li, H.; Teng, X.; Du, Y.; Miao, Y.; Lin, H.-C.; Han, D. Cancer diagnosis with DNA molecular computation. Nat. Nanotechnol. 2020, 15, 709–715. [Google Scholar] [CrossRef]
- Adleman, L.M. Molecular computation of solutions to combinatorial problems. Science 1994, 266, 1021. [Google Scholar] [CrossRef] [Green Version]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Tan, W. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res. 2010, 43, 48–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer Beacons for the Direct Detection of Proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831. [Google Scholar] [CrossRef]
- Sakai, Y.; Islam, M.S.; Adamiak, M.; Shiu, S.C.; Tanner, J.A.; Heddle, J.G. DNA Aptamers for the Functionalisation of DNA Origami Nanostructures. Genes 2018, 9, 571. [Google Scholar] [CrossRef] [Green Version]
- Lieblein, A.L.; Fürtig, B.; Schwalbe, H. Optimizing the Kinetics and Thermodynamics of DNA i-Motif Folding. ChemBioChem 2013, 14, 1226–1230. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, M.; Xing, Y.; Li, Y.; Joedecke, C.-C.; Jin, J.; Yang, Z.; Liu, D. Study of pH-Induced Folding and Unfolding Kinetics of the DNA i-Motif by Stopped-Flow Circular Dichroism. Langmuir 2012, 28, 17743–17748. [Google Scholar] [CrossRef] [PubMed]
- Ijäs, H.; Hakaste, I.; Shen, B.; Kostiainen, M.A.; Linko, V. Reconfigurable DNA Origami Nanocapsule for pH-Controlled Encapsulation and Display of Cargo. ACS Nano 2019, 13, 5959–5967. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Yang, Y.; Cheng, E.; Zhao, M.; Meng, H.; Liu, D.; Zhou, D. A pH-driven, reconfigurable DNA nanotriangle. Chem. Commun. 2009, 824–826. [Google Scholar] [CrossRef]
- Leung, K.; Chakraborty, K.; Saminathan, A.; Krishnan, Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 2019, 14, 176–183. [Google Scholar] [CrossRef]
- Li, T.; Famulok, M. I-Motif-Programmed Functionalization of DNA Nanocircles. J. Am. Chem. Soc. 2013, 135, 1593–1599. [Google Scholar] [CrossRef]
- Amodio, A.; Zhao, B.; Porchetta, A.; Idili, A.; Castronovo, M.; Fan, C.; Ricci, F. Rational Design of pH-Controlled DNA Strand Displacement. J. Am. Chem. Soc. 2014, 136, 16469–16472. [Google Scholar] [CrossRef] [PubMed]
- Idili, A.; Vallée-Bélisle, A.; Ricci, F. Programmable pH-Triggered DNA Nanoswitches. J. Am. Chem. Soc. 2014, 136, 5836–5839. [Google Scholar] [CrossRef] [PubMed]
- Kohman, R.E.; Han, X. Light sensitization of DNA nanostructures via incorporation of photo-cleavable spacers. Chem. Commun. 2015, 51, 5747–5750. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, Y.; Asanuma, H. Light-Driven DNA Nanomachine with a Photoresponsive Molecular Engine. Acc. Chem. Res. 2014, 47, 1663–1672. [Google Scholar] [CrossRef]
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science 2015, 347, 1446. [Google Scholar] [CrossRef]
- Arnott, P.M.; Howorka, S. A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport across Membranes. ACS Nano 2019, 13, 3334–3340. [Google Scholar] [CrossRef]
- Mao, C.; Sun, W.; Shen, Z.; Seeman, N.C. A nanomechanical device based on the B–Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar] [CrossRef]
- Marras, A.E.; Shi, Z.; Lindell, M.G.; Patton, R.A.; Huang, C.-M.; Zhou, L.; Su, H.-J.; Arya, G.; Castro, C.E. Cation-Activated Avidity for Rapid Reconfiguration of DNA Nanodevices. ACS Nano 2018, 12, 9484–9494. [Google Scholar] [CrossRef]
- Sannohe, Y.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Visualization of Dynamic Conformational Switching of the G-Quadruplex in a DNA Nanostructure. J. Am. Chem. Soc. 2010, 132, 16311–16313. [Google Scholar] [CrossRef]
- Tang, Q.; Lai, W.; Wang, P.; Xiong, X.; Xiao, M.; Li, L.; Fan, C.; Pei, H. Multi-Mode Reconfigurable DNA-Based Chemical Reaction Circuits for Soft Matter Computing and Control. Angew. Chem. Int. Ed. 2021, 60, 15013–15019. [Google Scholar] [CrossRef]
- Kuzyk, A.; Yang, Y.; Duan, X.; Stoll, S.; Govorov, A.O.; Sugiyama, H.; Endo, M.; Liu, N. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 2016, 7, 10591. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3, 103–113. [Google Scholar] [CrossRef]
- Wolter, O.; Mayer, G. Aptamers as Valuable Molecular Tools in Neurosciences. J. Neurosci. 2017, 37, 2517. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Glaeser, R.M.; Nogales, E. How Cryo-EM Became so Hot. Cell 2017, 171, 1229–1231. [Google Scholar] [CrossRef]
- Liu, Y.; Gonen, S.; Gonen, T.; Yeates, T.O. Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system. Proc. Natl. Acad. Sci. USA 2018, 115, 3362. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.-C.; Martin, T.G.; Scheres, S.H.W.; Dietz, H. Cryo-EM structure of a 3D DNA-origami object. Proc. Natl. Acad. Sci. USA 2012, 109, 20012. [Google Scholar] [CrossRef] [Green Version]
- Kube, M.; Kohler, F.; Feigl, E.; Nagel-Yüksel, B.; Willner, E.M.; Funke, J.J.; Gerling, T.; Stömmer, P.; Honemann, M.N.; Martin, T.G.; et al. Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution. Nat. Commun. 2020, 11, 6229. [Google Scholar] [CrossRef]
- Bertosin, E.; Stömmer, P.; Feigl, E.; Wenig, M.; Honemann, M.N.; Dietz, H. Cryo-Electron Microscopy and Mass Analysis of Oligolysine-Coated DNA Nanostructures. ACS Nano 2021, 15, 9391–9403. [Google Scholar] [CrossRef]
- Lyumkis, D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 2019, 294, 5181–5197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, E. The development of cryo-EM into a mainstream structural biology technique. Nat. Methods 2016, 13, 24–27. [Google Scholar] [CrossRef]
- Carragher, B.; Cheng, Y.; Frost, A.; Glaeser, R.M.; Lander, G.C.; Nogales, E.; Wang, H.W. Current outcomes when optimizing ‘standard’ sample preparation for single-particle cryo-EM. J. Microsc. 2019, 276, 39–45. [Google Scholar] [CrossRef]
- Aksel, T.; Yu, Z.; Cheng, Y.; Douglas, S.M. Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Nat. Biotechnol. 2021, 39, 378–386. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, S.; Zhang, S.; Sodroski, J.; Yang, Z.; Liu, D.; Mao, Y. Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins. Angew. Chem. Int. Ed. 2018, 57, 2072–2076. [Google Scholar] [CrossRef]
- Selmi, D.N.; Adamson, R.J.; Attrill, H.; Goddard, A.D.; Gilbert, R.J.C.; Watts, A.; Turberfield, A.J. DNA-Templated Protein Arrays for Single-Molecule Imaging. Nano Lett. 2011, 11, 657–660. [Google Scholar] [CrossRef]
- Aissaoui, N.; Lai-Kee-Him, J.; Mills, A.; Declerck, N.; Morichaud, Z.; Brodolin, K.; Baconnais, S.; Le Cam, E.; Charbonnier, J.B.; Sounier, R.; et al. Modular Imaging Scaffold for Single-Particle Electron Microscopy. ACS Nano 2021, 15, 4186–4196. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, M.; Hogle, J.M.; Shih, W.M.; Wagner, G.; Nasr, M.L. DNA-Corralled Nanodiscs for the Structural and Functional Characterization of Membrane Proteins and Viral Entry. J. Am. Chem. Soc. 2018, 140, 10639–10643. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.A.R.; Lund, K.; Liu, Y.; Yan, H.; Chaput, J.C. Self-Assembled Peptide Nanoarrays: An Approach to Studying Protein–Protein Interactions. Angew. Chem. Int. Ed. 2007, 46, 3051–3054. [Google Scholar] [CrossRef]
- Rosier, B.J.H.M.; Markvoort, A.J.; Gumí Audenis, B.; Roodhuizen, J.A.L.; den Hamer, A.; Brunsveld, L.; de Greef, T.F.A. Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome. Nat. Catal. 2020, 3, 295–306. [Google Scholar] [CrossRef]
- Kostrz, D.; Wayment-Steele, H.K.; Wang, J.L.; Follenfant, M.; Pande, V.S.; Strick, T.R.; Gosse, C. A modular DNA scaffold to study protein–protein interactions at single-molecule resolution. Nat. Nanotechnol. 2019, 14, 988–993. [Google Scholar] [CrossRef]
- Shaw, A.; Hoffecker, I.T.; Smyrlaki, I.; Rosa, J.; Grevys, A.; Bratlie, D.; Sandlie, I.; Michaelsen, T.E.; Andersen, J.T.; Högberg, B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 2019, 14, 184–190. [Google Scholar] [CrossRef]
- Veneziano, R.; Moyer, T.J.; Stone, M.B.; Wamhoff, E.-C.; Read, B.J.; Mukherjee, S.; Shepherd, T.R.; Das, J.; Schief, W.R.; Irvine, D.J.; et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 2020, 15, 716–723. [Google Scholar] [CrossRef]
- Iwaki, M.; Wickham, S.F.; Ikezaki, K.; Yanagida, T.; Shih, W.M. A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads. Nat. Commun. 2016, 7, 13715. [Google Scholar] [CrossRef] [Green Version]
- Funke, J.J.; Ketterer, P.; Lieleg, C.; Schunter, S.; Korber, P.; Dietz, H. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2016, 2, e1600974. [Google Scholar] [CrossRef] [Green Version]
- Nickels, P.C.; Wünsch, B.; Holzmeister, P.; Bae, W.; Kneer, L.M.; Grohmann, D.; Tinnefeld, P.; Liedl, T. Molecular force spectroscopy with a DNA origami–based nanoscopic force clamp. Science 2016, 354, 305–307. [Google Scholar] [CrossRef]
- Blakely, B.L.; Dumelin, C.E.; Trappmann, B.; McGregor, L.M.; Choi, C.K.; Anthony, P.C.; Duesterberg, V.K.; Baker, B.M.; Block, S.M.; Liu, D.R.; et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat. Methods 2014, 11, 1229–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, F.; Li, M.; Mao, X.; Li, F.; Xiang, X.; Li, Q.; Wang, L.; Zuo, X.; Fan, C.; Zhu, Y. DNA Framework-Based Topological Cell Sorters. Angew. Chem. Int. Ed. 2020, 59, 10406–10410. [Google Scholar] [CrossRef]
- Zhang, Y.; Ge, C.; Zhu, C.; Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 2014, 5, 5167. [Google Scholar] [CrossRef]
- Dutta, P.K.; Zhang, Y.; Blanchard, A.T.; Ge, C.; Rushdi, M.; Weiss, K.; Zhu, C.; Ke, Y.; Salaita, K. Programmable Multivalent DNA-Origami Tension Probes for Reporting Cellular Traction Forces. Nano Lett. 2018, 18, 4803–4811. [Google Scholar] [CrossRef]
- Brockman, J.M.; Blanchard, A.T.; Pui-Yan, V.; Derricotte, W.D.; Zhang, Y.; Fay, M.E.; Lam, W.A.; Evangelista, F.A.; Mattheyses, A.L.; Salaita, K. Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 2018, 15, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Glazier, R.; Brockman, J.M.; Bartle, E.; Mattheyses, A.L.; Destaing, O.; Salaita, K. DNA mechanotechnology reveals that integrin receptors apply pN forces in podosomes on fluid substrates. Nat. Commun. 2019, 10, 4507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, R.; Kellner, A.V.; Ma, V.P.Y.; Su, H.; Deal, B.R.; Brockman, J.M.; Salaita, K. DNA probes that store mechanical information reveal transient piconewton forces applied by T cells. Proc. Natl. Acad. Sci. USA 2019, 116, 16949–16954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, M.C.; Smith, D.M.; Jobst, M.A.; Sajfutdinow, M.; Liedl, T.; Romano, F.; Rovigatti, L.; Louis, A.A.; Doye, J.P.K. Force-Induced Unravelling of DNA Origami. ACS Nano 2018, 12, 6734–6747. [Google Scholar] [CrossRef]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef] [PubMed]
- Gerling, T.; Kube, M.; Kick, B.; Dietz, H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018, 4, eaau1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastassacos, F.M.; Zhao, Z.; Zeng, Y.; Shih, W.M. Glutaraldehyde Cross-Linking of Oligolysines Coating DNA Origami Greatly Reduces Susceptibility to Nuclease Degradation. J. Am. Chem. Soc. 2020, 142, 3311–3315. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R. Nuclease resistance of DNA nanostructures. Nat. Rev. Chem. 2021, 5, 225–239. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Han, D. Obtaining Precise Molecular Information via DNA Nanotechnology. Membranes 2021, 11, 683. https://doi.org/10.3390/membranes11090683
Tang Q, Han D. Obtaining Precise Molecular Information via DNA Nanotechnology. Membranes. 2021; 11(9):683. https://doi.org/10.3390/membranes11090683
Chicago/Turabian StyleTang, Qian, and Da Han. 2021. "Obtaining Precise Molecular Information via DNA Nanotechnology" Membranes 11, no. 9: 683. https://doi.org/10.3390/membranes11090683
APA StyleTang, Q., & Han, D. (2021). Obtaining Precise Molecular Information via DNA Nanotechnology. Membranes, 11(9), 683. https://doi.org/10.3390/membranes11090683





