Homo Sapiens (Hsa)-microRNA (miR)-6727-5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture and Maintenance of Human Neonatal Keratinocytes and Fibroblasts and Human Adult Fibroblasts
2.2. Efflux of [3H]Cholesterol to HDL from Human Keratinocytes and Fibroblasts
2.3. In Vitro Wound Healing: Scrape (‘Scratch’) Wound Assays
2.4. Total RNA Isolation, Microchip Analysis and Quantitative PCR
2.5. Statistical Analyses
3. Results
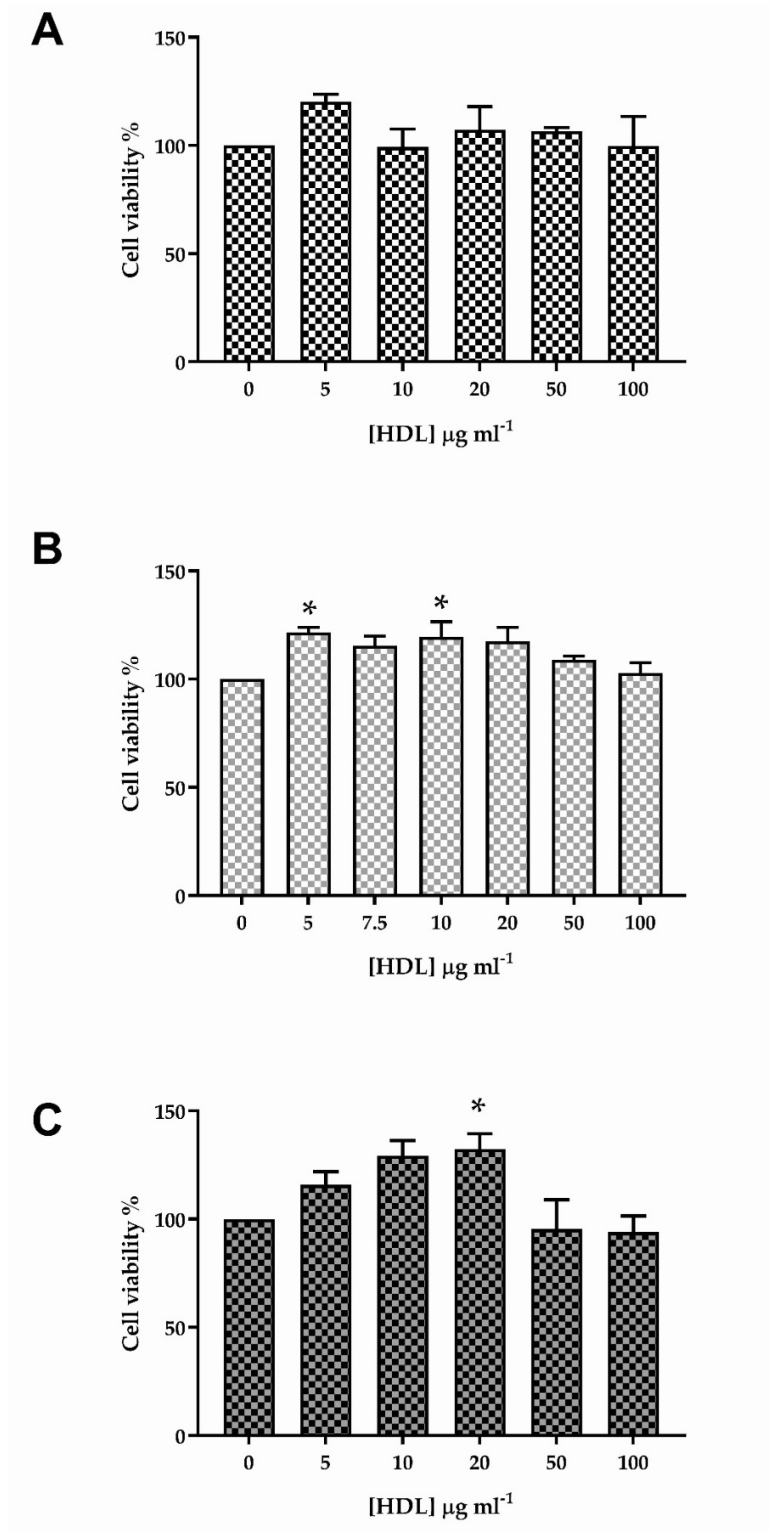
3.1. The Effect of HDL on Viability of HKn, HDFn and HDFa
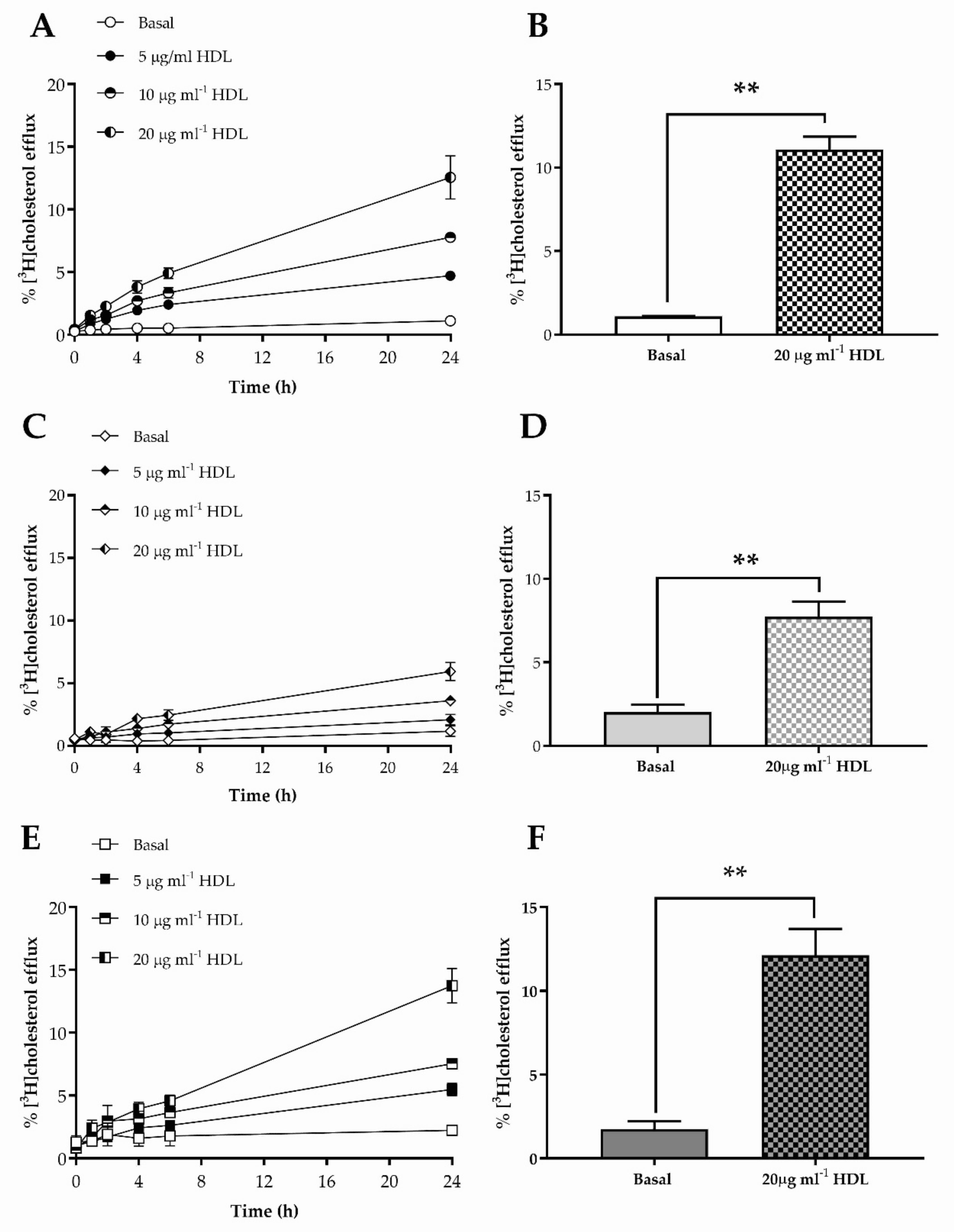
3.2. Cholesterol Efflux to HDL from HKn, HDFn and HDFa
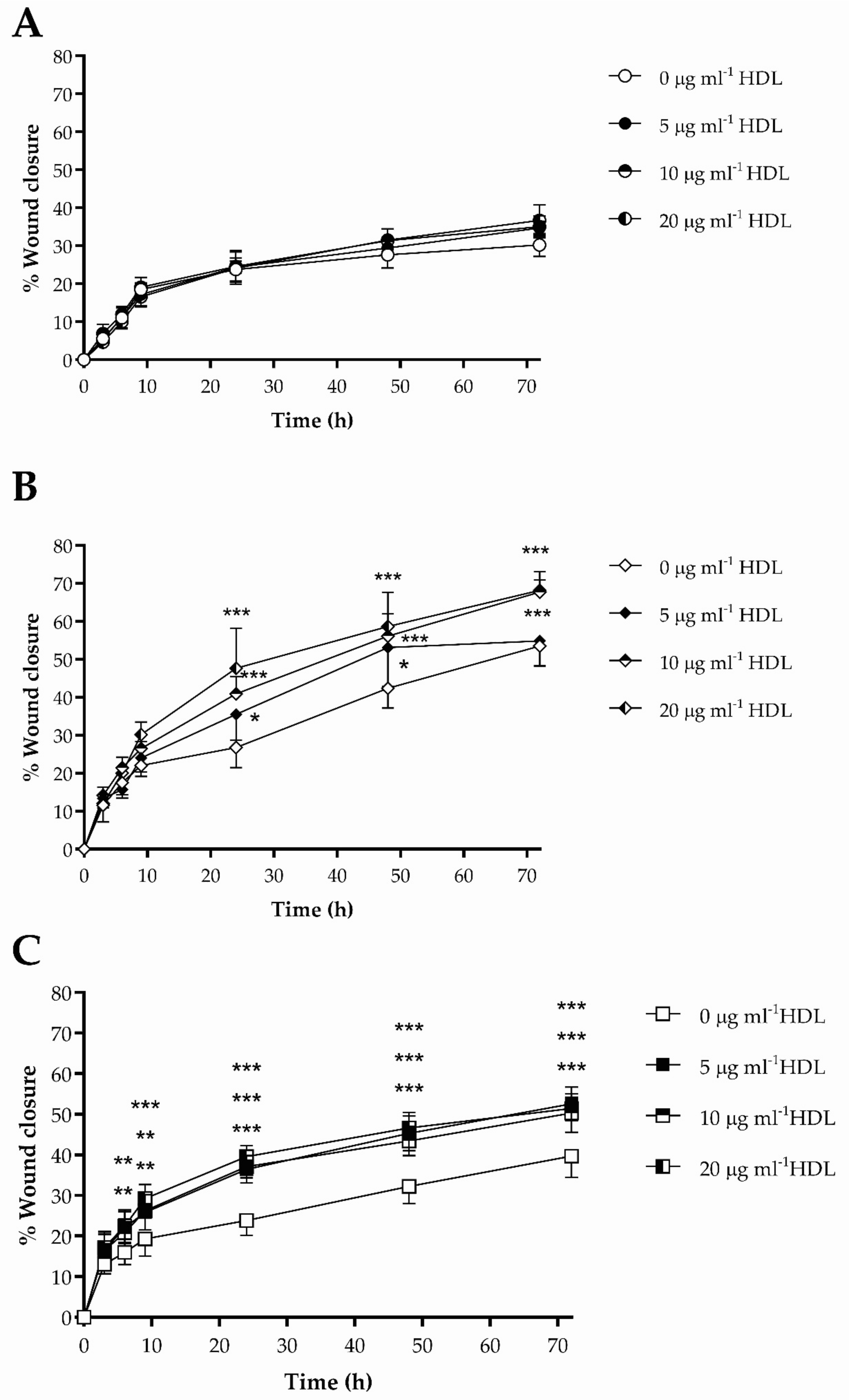
3.3. The Impact of HDL on Scrape Wound Healing by HKn, HDFn and HDFa
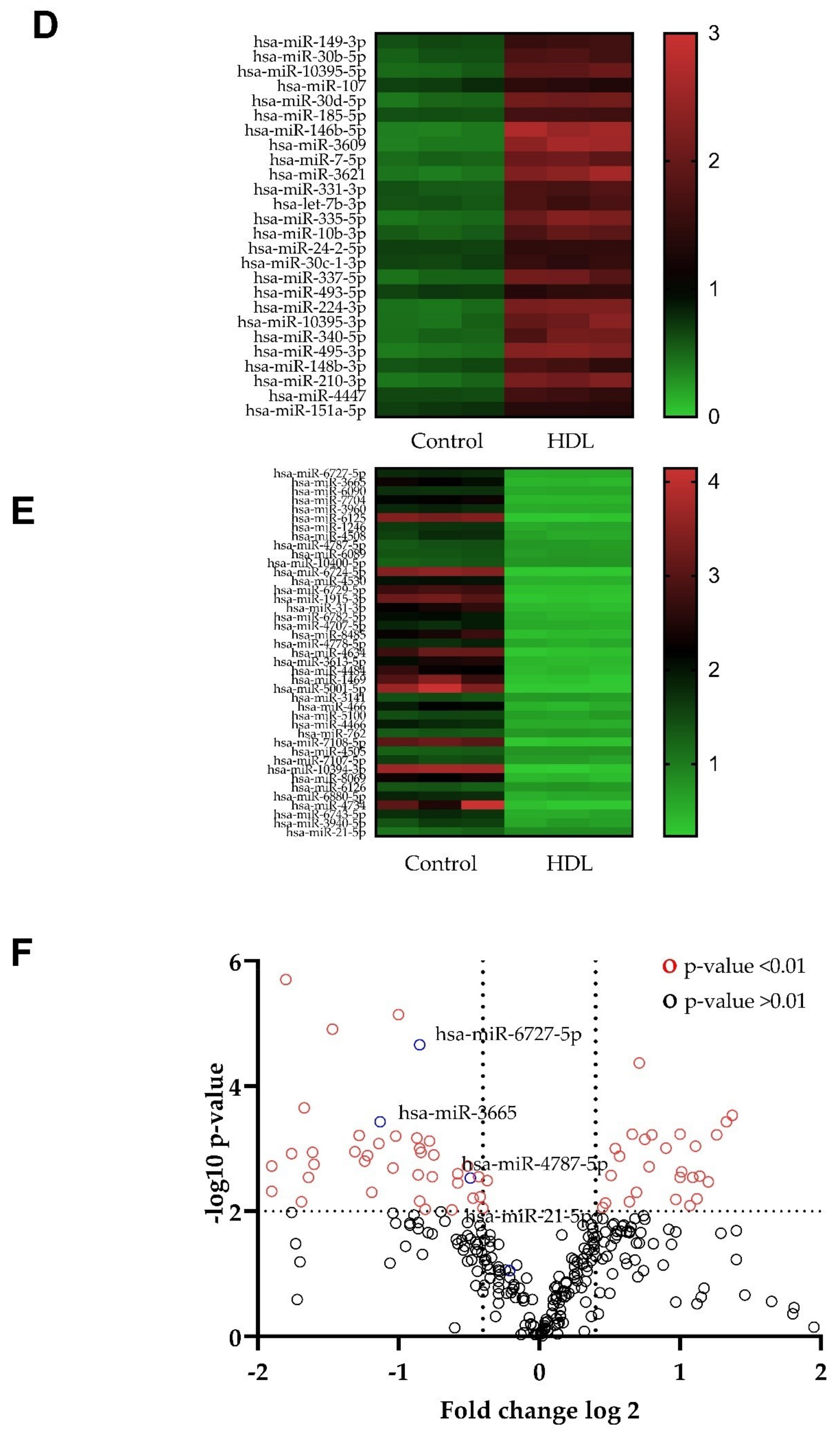
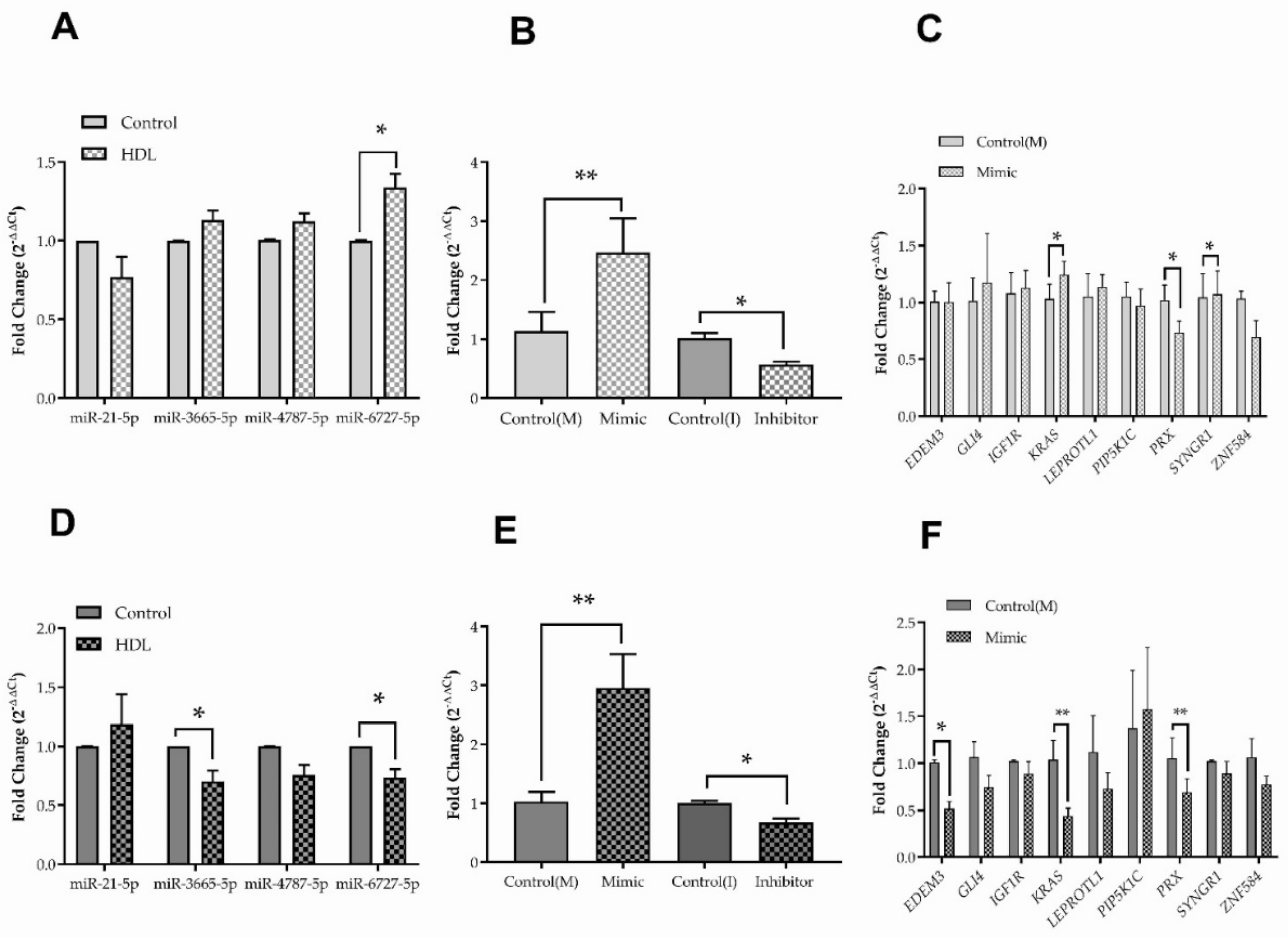
3.4. Changes in microRNA Expression Caused by Exposure to HDL in HDFn and HDFa
3.5. Hsa-miR-6727-5p: Regulation by HDL, Delivery and Efficacy
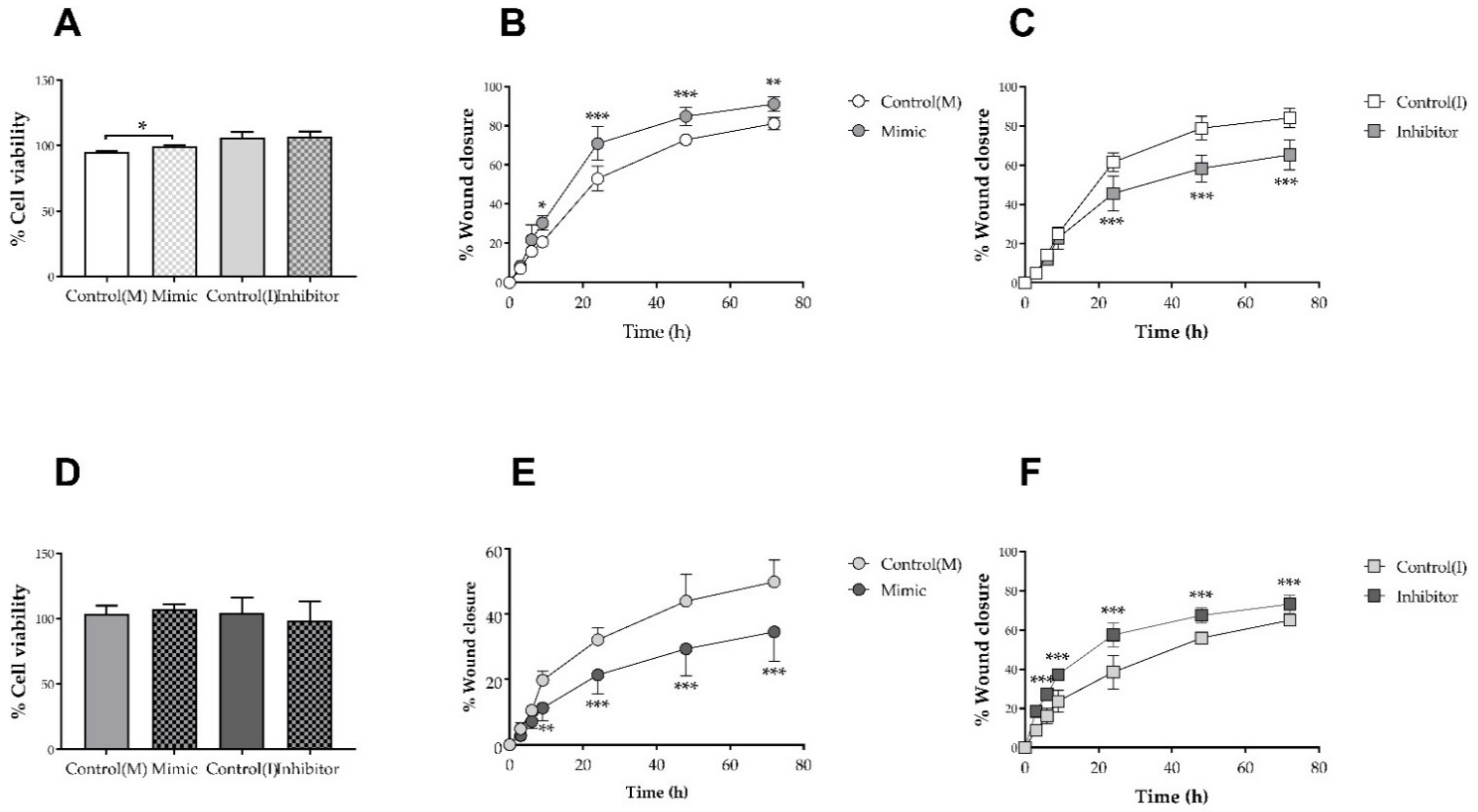
3.6. Hsa-miR-6727-5p Mimic and Inhibitor: Impact on Cell Viability and Scrape Wound Healing in HDFn and HDFa
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Guest, J.F.; Fuller, F.W.; Vowden, P. Cohort study evaluating the burden of wounds to the UK’s National Health Service in 2017/2018: Update from 2012/2013. BMJ Open 2020, 10, e045253. [Google Scholar] [CrossRef] [PubMed]
- Golebiewski, E.M.; Poole, W.M. Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev. 2015, 29, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielefeld, K.A.; Amini-Nik, S.; Alman, B.A. Cutaneous wound healing: Recruiting developmental pathways for regeneration. Cell. Mol. Life Sci. 2013, 70, 2059–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, P.; Nunan, R. Cellular and molecular mechanisms of repair in acute and chronic wound healing. Br. J. Dermatol. 2015, 173, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Wicks, K.; Torbica, T.; Mace, K.A. Myeloid cell dysfunction and the pathogenesis of the diabetic chronic wound. Semin. Immunol. 2014, 26, 341–353. [Google Scholar] [CrossRef]
- Schneider, C.; Stratman, S.; Kirsner, R.S. Lower Extremity Ulcers. Med. Clin. N. Am. 2021, 105, 663–679. [Google Scholar] [CrossRef]
- Süntar, I.; Çetinkaya, S.; Panieri, E.; Saha, S.; Buttari, B.; Profumo, E.; Saso, L. Regulatory Role of Nrf2 Signaling Pathway in Wound Healing Process. Molecules 2021, 26, 2424. [Google Scholar] [CrossRef]
- Versey, Z.; da Cruz Nizer, W.S.; Russell, E.; Zigic, S.; DeZeeuw, K.G.; Marek, J.E.; Overhage, J.; Cassol, E. Biofilm-Innate Immune Interface: Contribution to Chronic Wound Formation. Front. Immunol. 2021, 12, 648554. [Google Scholar] [CrossRef]
- Takematsu, E.; Spencer, A.; Auster, J.; Chen, P.-C.; Graham, A.; Martin, P.; Baker, A.B. Genome wide analysis of gene expression changes in skin from patients with type 2 diabetes. PLoS ONE 2020, 15, e0225267. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.Y.; Marinkovich, M.P.; Lucas, E.; Gorell, E.; Chiou, A.; Lu, Y.; Gillon, J.; Patel, D.; Rudin, D. A systematic literature review of the disease burden in patients with recessive dystrophic epidermolysis bullosa. Orphanet J. Rare Dis. 2021, 16, 175. [Google Scholar] [CrossRef]
- Sen, C.K. Human Wound and Its Burden: Updated 2020 Compendium of Estimates. Adv. Wound Care 2021, 10, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Gordts, S.C.; Muthuramu, I.; Amin, R.; Jacobs, F.; De Geest, B. The Impact of Lipoproteins on Wound Healing: Topical HDL Therapy Corrects Delayed Wound Healing in Apolipoprotein E Deficient Mice. Pharmaceuticals 2014, 7, 419–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Yin, H.; Liu, P.; Zhang, H.; She, M. Essential role of HDL on endothelial progenitor cell proliferation with PI3K/Akt/cyclin D1 as the signal pathway. Exp. Biol. Med. 2010, 235, 1082–1092. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.T.M.; Prosser, H.C.G.; Dunn, L.L.; Vanags, L.Z.; Ridiandries, A.; Tsatralis, T.; Lecce, L.; Clayton, Z.E.; Yuen, S.C.G.; Roberston, S.; et al. High-density lipoproteins rescue diabetes-impaired angiogenesis via scavenger receptor class B type I. Diabetes 2016, 65, 3091–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsatralis, T.; Ridiandries, A.; Roberson, S.; Vanages, L.Z.; Lam, Y.T.; Tan, J.M.T.; Ng, M.K.C.; Bursill, C.A. Reconstituted high-density lipoproteins promote wound repair and blood flow recovery in response to ischemia in aged mice. Lipids Health Dis. 2016, 15, 150. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.; Yu, B.; Ren, H.; Willard, B.; Pan, L.; Zu, L.; Shen, X.; Ma, Y.; Li, X.; Nui, C.; et al. High-density lipoprotein nitration and chlorination catalyzed by myeloperoxidase impair its effect of promoting endothelial repair. Free Radic. Biol. Med. 2013, 60, 272–281. [Google Scholar] [CrossRef]
- Speer, T.; Rohrer, L.; Blysszczuk, P.; Shroff, R.; Kuschnerus, K.; Krankel, N.; Kania, G.; Zewinger, S.; Akhmedov, A.; Shi, Y.; et al. Abnormal High-Density Lipoprotein Induces Endothelial Dysfunction via Activation of Toll-like Receptor-2. Immunity 2013, 38, 754–768. [Google Scholar] [CrossRef] [Green Version]
- Lv, P.; Tong, X.; Peng, Q.; Liu, Y.; Jin, H.; Liu, R.; Sun, W.; Pan, B.; Zheng, L.; Huang, Y. Treatment with the herbal medicine, nanoxintong improves the protective effect of high-density lipoproteins on endothelial function in patients with type 2 diabetes. Mol. Med. Rep. 2016, 13, 2007–2016. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhao, M.; He, D.; Zhao, X.; Zhang, W.; Wei, L.; Huang, E.; Ji, L.; Zhang, M.; Willard, B.; et al. HDL in diabetic nephropathy has less effect in diabetic repairing than diabetes without complications. Lipids Health Dis. 2016, 15, 76. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, M.A.; Chauhuri, P.; Abelson, B.; Cross, B.N.; Graham, L.M. Apolipoprotein A-I mimetic peptide reverses impaired arterial healing after injury by reducing oxidative stress. Atherosclerosis 2015, 241, 709–715. [Google Scholar] [CrossRef] [Green Version]
- He, D.; Zhao, M.; Wu, C.; Zhang, W.; Niu, C.; Yu, B.; Jin, J.; Ji, L.; Willard, B.; Mathew, A.; et al. Apolipoprotein A-I mimietic peptide 4F promotes endothelial repairing and compromises reendothelialization impaired by oxidized HDL through SR-B1. Redox Biol. 2018, 15, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Qian, M.; Huang, C.; Cui, P.; Li, W.; Du, Q.; Yi, S.; Shi, X.; Guo, Y.; Zheng, J.; et al. Comparison of Mechanisms of Endothelial Cell Protections Between High-Density Lipoprotein and Apolipoprotein A-I Mimetic Peptide. Front. Pharmacol. 2019, 10, 817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Jin, J.; Wang, Y.; Sun, J. High density lipoprotein promoting proliferation and migration of type II alveolar epithelial cells during inflammation state. Lipids Health Dis. 2018, 16, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Calvert, A.E.; Kaplan, N.; McMahon, K.M.; Yang, W.; Lu, K.Q.; Peng, H.; Thaxton, C.S.; Lavker, R.M. HDL Nanoparticles Have Wound Healing and Anti-Inflammatory Properties and Can Topically Deliver miRNA. Adv. Ther. 2020, 3, 2000138. [Google Scholar] [CrossRef]
- Vickers, K.C.; Michell, D.L. HDL-small RNA Export, Transport, and Functional Delivery in Atherosclerosis. Curr. Atheroscler. Rep. 2021, 23, 38. [Google Scholar] [CrossRef]
- Kajani, S.; Curley, S.; McGillicuddy, F.C. Unravelling HDL—Looking beyond the Cholesterol Surface to the Quality Within. Int. J. Mol. Sci. 2018, 19, 1971. [Google Scholar] [CrossRef] [Green Version]
- Aryl, B.; Singh, A.K.; Rotllan, N.; Price, N.; Fernandez-Hernando, C. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef]
- Tarlton, J.M.R.; Lightbody, R.J.; Patterson, S.; Graham, A. Protection against Glucolipotoxicity by High Density Lipoprotein in Human PANC-1 Hybrid 1.1B4 Pancreatic Beta Cells: The Role of microRNA. Biology 2021, 10, 218. [Google Scholar] [CrossRef]
- Wang, F.; Gao, Y.; Yuan, Y.; Du, R.; Li, P.; Liu, F.; Tian, Y.; Wang, Y.; Zhang, R.; Zhao, B.; et al. MicroRNA-31 Can Positively Regulate the Proliferation, Differentiation and Migration of Keratinocytes. Biomed. Hub 2020, 5, 93–104. [Google Scholar] [CrossRef]
- Niemiec, S.M.; Louiselle, A.E.; Liechty, K.W.; Zgheib, C. Role of microRNA in Pressure Ulcer Immune Response, Pathogenesis, and Treatment. Int. J. Mol. Sci. 2020, 22, 64. [Google Scholar] [CrossRef]
- Petkovic, M.; Sorensen, A.E.; Leal, E.C.; Carvalho, E.; Dalgaard, L.T. Mechanistic Actions of microRNAs in Diabetic Wound Healing. Cells 2020, 9, 2228. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Zhao, J.; Ling, H.; Deng, Y.; Li, X.; He, Y. Exploring microRNAs in diabetic chronic cutaneous ulcers: Regulatory mechanisms and therapeutic potential. J. Cereb. Blood Flow Metab. 2020, 177, 4077–4095. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Horobin, R.W.; Colombo, L.L.; Blazquez-Castro, A. Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018, 120, 159–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.M.; Allen, A.M.; Graham, A. Targeting mitochondrial 18 kDa translocator protein (TSPO) regulates macrophage cholesterol efflux and lipid phenotype. Clin. Sci. 2014, 127, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Weibel, G.L.; Drazul-Schrader, D.; Shivers, D.L.; Wade, A.N.; Rothblat, G.H.; Reilly, M.P.; de la Llera-Moya, M. Importance of Evaluating Cell Cholesterol Influx with Efflux in Determining the Impact of Human Serum on Cholesterol Metabolism and Atherosclerosis. Arter. Thromb. Vasc. Biol. 2014, 34, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Llera Moya, M.; Atger, V.; Paul, J.L.; Fournier, N.; Moatti, N.; Giral, P.; Friday, K.E.; Rothblat, G. A cell culture system for screening human serum for ability to promote cellular cholesterol efflux. Relations between serum components and efflux, esterification, and transfer. Arter. Thromb. Vasc. Biol. 1994, 14, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Faniku, C.; O’Shaughnessy, E.; Lorraine, C.; Johnstone, S.R.; Graham, A.; Greenhough, S.; Martin, P.E.M. The Connexin Mimetic Peptide Gap27 and Cx43-Knockdown Reveal Differential Roles for Connexin43 in Wound Closure Events in Skin Model Systems. Int. J. Mol. Sci. 2018, 19, 604. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Schmitten, R.D.; Livaks, K.J. Analysing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, W.J. Volcano plots in analysing differential expressions with mRNA microarrays. J. Bioinform. Comput. Biol. 2012, 10, 1231003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.J.; Biao, L.; Kim, P.; Elias, P.M.; Feingold, K.R. Regulation of ABCA1 expression in human keratinocytes and murine epidermis. J. Lipid Res. 2006, 47, 2248–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.J.; Biao, L.; Tarling, E.J.; Kim, P.; Man, M.-Q.; Crumrine, D.; Edwards, P.A.; Elias, P.M.; Feingold, K.R. Regulation of ABCG1 expression in human keratinocytes and murine epidermis. J. Lipid Res. 2010, 51, 3185–3195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picardo, M.; Massey, J.B.; Kuhn, D.E.; Gotto, A.M., Jr.; Gianturco, S.H.; Pownall, H.J. Partially reassembled high density lipoproteins. Effects on cholesterol flux, synthesis, and esterification in normal human skin fibroblasts. Arteriosclerosis 1986, 6, 434–441. [Google Scholar] [CrossRef] [Green Version]
- Marcil, M.; Yu, L.; Krimbou, L.; Boucher, B.; Oram, J.F.; Cohn, J.S.; Genest, J., Jr. Cellular Cholesterol Transport and Efflux in Fibroblasts Are Abnormal in Subjects with Familial HDL Deficiency. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 159–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Jin, R.; Qi, Y.; Zhou, H.; Zhu, T.; Liu, L.; Gu, Y.; Luan, K.; Luo, X.; Zhang, S. Cholesterol partially rescues the inhibition effect of pravastatin on keratinocytes proliferation by regulating cell cycle relative proteins through AKT and ERK pathway. Dermatol. Ther. 2020, 33, e14305. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Latif, M.I.A.; Murota, H.; Terao, M.; Katayama, I. Effects of a 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor and low-density lipoprotein on proliferation and migration of keratinocytes. Br. J. Dermatol. 2010, 163, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Neuwirt, H.; Bouchal, J.; Kharaishvili, G.; Ploner, C.; Johrer, K.; Pitterl, F.; Weber, A.; Klocker, H.; Eder, I.E. Cancer-associated fibroblasts promote prostate tumor growth and progression through upregulation of cholesterol and steroid biosynthesis. Cell Commun. Signal. 2020, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- Ostlund, R.E., Jr.; Yang, Y.W. Effect of cholesterol and growth factors on the proliferation of cultured human skin fibroblasts. Exp. Cell Res. 1985, 161, 509–516. [Google Scholar] [CrossRef]
- Mateu, R.; Zivicova, V.; Krejci, E.D.; Grim, M.; Strnad, H.; Vlcek, C.; Kolar, M.; Lacina, L.; Gal, P.; Borsky, J.; et al. Functional differences between neonatal and adult fibroblasts and keratinocytes: Donor age affects epithelial-mesenchymal crosstalk in vitro. Int. J. Mol. Med. 2016, 38, 1063–1074. [Google Scholar] [CrossRef] [Green Version]
- Desai, R.; Frazier, A.E.; Durigon, R.; Patel, H.; Jones, A.W.; Dalla Rosa, I.; Lake, N.J.; Compton, A.G.; Mountford, H.S.; Tucker, E.J.; et al. ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain 2017, 140, 1595–1610. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Dana, S.E.; Brown, M.S. Esterification of Low Density Lipoprotein Cholesterol in Human Fibroblasts and Its Absence in Homozygous Familial Hypercholesterolemia. Proc. Natl. Acad. Sci. USA 1974, 71, 4288–4292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunniff, C.; Kratz, L.E.; Moser, A.; Natowicz, M.R.; Kelley, R.I. Clinical and biochemical spectrum of patients with RSH/Smith-Lemli-Opitz syndrome and abnormal cholesterol metabolism. Am. J. Med. Genet. 1997, 68, 236–269. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kondo, H.; Shimada, Y.; Waheed, A.A.; Ohno-Iwashita, Y. Cellular aging-dependent decrease in cholesterol in membrane microdomains of human diploid fibroblasts. Exp. Cell Res. 2003, 290, 381–390. [Google Scholar] [CrossRef]
- Ethier, M.F.; Hickler, R.B.; Saunders, R.H. Cholesterol and cholesteryl ester concentration in different size classes of cultured human fibroblasts: Effect of in vitro aging. Mech. Ageing Dev. 1982, 20, 165–174. [Google Scholar] [CrossRef]
- Wang, B.; Tontonoz, P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 2018, 14, 452–463. [Google Scholar] [CrossRef]
- Jensen, S.G.; Lamy, P.; Rasmussen, M.D.; Ostenfeld, M.S.; Dysrkjot, L.; Orntoft, T.F.; Andersen, C.L. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genom. 2011, 12, 435. [Google Scholar] [CrossRef] [Green Version]
- Lunn, M.L.; Mouritzen, P.; Faber, K.; Jacobsen, N. MicroRNA quantitation from a single cell by PCR using SYBR® Green detection and LNA-based primers. Nat. Chem. Biol. 2008, 5, iii–iv. [Google Scholar] [CrossRef]
- Ballantyne, K.N.; van Oorschot, R.A.H.; Mitchell, R.J. Locked nucleic acids in PCR primers increase sensitivity and performance. Genomics 2008, 91, 301–305. [Google Scholar] [CrossRef] [Green Version]
- Busk, P.K. A tool for design of primers for microRNA-specific quantitative RT-qPCR’. BMC Bioinform. 2014, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Pei, G.; Song, M.; Dai, S.; Wang, Y. Influence of hsa-miR-6727-5p on the proliferation, apoptosis, invasion and migration of Caski, Hela and SiHa cervical cell lines. J. BUON 2017, 22, 974–978. [Google Scholar] [PubMed]
- Tey, S.; Shahrizail, N.; Drew, A.P.; Samulong, S.; Goh, K.-J.; Battaloglu, E.; Atkinson, D.; Parman, Y.; Jordanova, A.; Chung, K.W.; et al. Linkage analysis and whole exome sequencing reveals AHNAK2 as a novel genetic cause for autosomal recessive CMT in a Malaysian family. Neurogenetics 2019, 20, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, D.B.; Bhaskara, A.; Droggiti, A.; Dickinson, S.; D’Antonio, M.; Mirsky, R.; Jessen, K.R. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J. Cell Biol. 2004, 164, 385–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, A.C.; Brophy, P.J. The function of the Periaxin gene during nerve repair in a model of CMT4F. J. Anat. 2002, 200, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Kerr, E.M.; Gaude, E.; Turrell, F.K.; Frezza, C.; Martins, C.P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nat. Cell Biol. 2016, 531, 110–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, E.; Weissmueller, S.; Morris, J.P., 4th; Chen, C.-C.; Wullenkord, R.; Lujambio, A.; de Stanchina, E.; Poirier, J.T.; Gainor, J.F.; Corcoran, R.B.; et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016, 534, 647–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polla, D.L.; Edmondson, A.C.; Duvet, S.; March, M.E.; Sousa, A.B.; Lehman, A.; Niyazov, D.; van Dijk, F.; Demirdas, S.; van Slegtenhorst, M.A.; et al. Bi-allelic variants in the ER quality-control mannosidase gene EDEM3 cause a congenital disorder of glycosylation. Am. J. Hum. Genet. 2021, 108, 1342–1439. [Google Scholar] [CrossRef]

| MiRs Primer | Source | miRbase Accession Number | Forward Primer Sequence 5′-3′ | Reverse Primer Sequence 5′-3′ |
|---|---|---|---|---|
| miR-1-3p | Human | MIMAT0000416 | UGGAAUGUAAAGAAGUAUGUAU | - |
| miR-6727-5p | Human | MIMAT0027355 | CAGGCGGCTGGGA | - |
| miR-3665 | Human | MIMTA0018087 | GAGCAGGTGCGGG | - |
| miR-4787-5p | Human | MIMAT0019956 | TGGCGGCGGCA OR GCGGGGGTGGCG | - |
| miR-21-5p | Human | NR_029493.1 | GCAGTAGCTTATCAGACTGATG | - |
| Gene Primer | Source | NCBI Reference Sequence | Forward Primer Sequence 5′-3′ | Reverse Primer Sequence 5′-3′ |
| RNU6 | Human | NR-104084.1 | TGACACGCAAATTCGTGAAG | - |
| SNORD-72 | Human | NR-002583 (80) | Product code: Hs_SNORD72_11 miScript Primer Assay, Cat. No.: 218300, GeneGlobe ID.: MS00033719 | |
| YWHAZ | Human | NM-001135701.2 | ACCGTTACTTGGCTGAGGTTGC | CCCAGTCTGATAGGATGTGTTGG |
| GAPDH | Human | - | Product code: Hs_GAPDH_1_SG, Cat. no.:249900 GeneGlobe ID.: QT00079247 | |
| RPL13α | Human | NM-012423.4 | CTCAAGGTGTTTGACGGCATCC | TACTTCCAGCCAACCTCGTGAG |
| KRAS | Human | NM-004985 | GGACTGGGGAGGGCTTTCT | GCCTGTTTTGTGTCTACTGTTCT |
| EDEM3 | Human | NM-025191 | CTCCCTGGCAATCATGCTGAA | AAGCCGGTAAAAGTTTGTAACCT |
| PIP5K1C | Human | NM-012398 | AGACCGTCATGCACAAGGAG | CAGTACAGCCCATAGAACTTGG |
| ZNF584 | Human | NM-173548 | GAGGCTCAGTTGGACCCATC | ACATCCCGGTATAGGCCCTTC |
| IGF1R | Human | NM-000875 | ATGCTGACCTCTGTTACCTCT | GGCTTATTCCCCACAATGTAGTT |
| GLI4 | Human | NM-138465 | GGAGTCAACTTCGGTCGGAG | TCAGGAGCAGCGAGTTATACT |
| LEPROTL1 | Human | NM-015344 | TTTGATGCTTGGATGTGCCCT | GCCCGTTGTAAGAAAGATGGC |
| SYNGR1 | Human | NM-004711 | GTGTTCGGCTCCATCGTGAA | GTTGGGGTTGCGGTTGTAGAT |
| PRX | Human | NM-181882 | GGTGGAAATTATCGTGGAGACG | GCAGCTCCCGAACGAAGAT |
| Universal Reverse Primer | Human | - | - | GAATCGAGCACCAGTTACGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastaki, K.M.; Tarlton, J.M.R.; Lightbody, R.J.; Graham, A.; Martin, P.E. Homo Sapiens (Hsa)-microRNA (miR)-6727-5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro. Membranes 2022, 12, 154. https://doi.org/10.3390/membranes12020154
Bastaki KM, Tarlton JMR, Lightbody RJ, Graham A, Martin PE. Homo Sapiens (Hsa)-microRNA (miR)-6727-5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro. Membranes. 2022; 12(2):154. https://doi.org/10.3390/membranes12020154
Chicago/Turabian StyleBastaki, Khaled Mahmoud, Jamie Maurice Roy Tarlton, Richard James Lightbody, Annette Graham, and Patricia Esther Martin. 2022. "Homo Sapiens (Hsa)-microRNA (miR)-6727-5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro" Membranes 12, no. 2: 154. https://doi.org/10.3390/membranes12020154
APA StyleBastaki, K. M., Tarlton, J. M. R., Lightbody, R. J., Graham, A., & Martin, P. E. (2022). Homo Sapiens (Hsa)-microRNA (miR)-6727-5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro. Membranes, 12(2), 154. https://doi.org/10.3390/membranes12020154







