Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence
Abstract
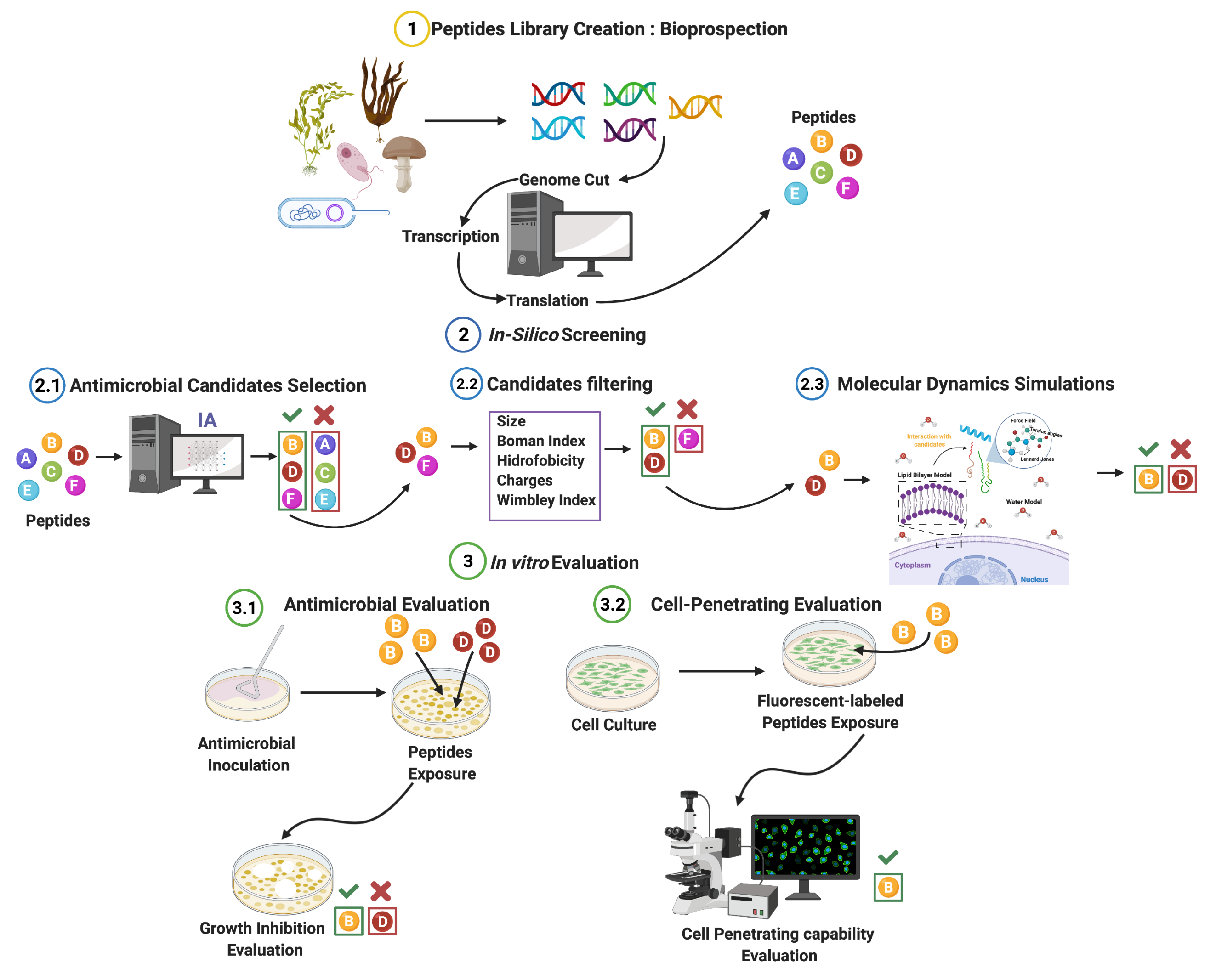
:1. Introduction
2. Materials and Methods
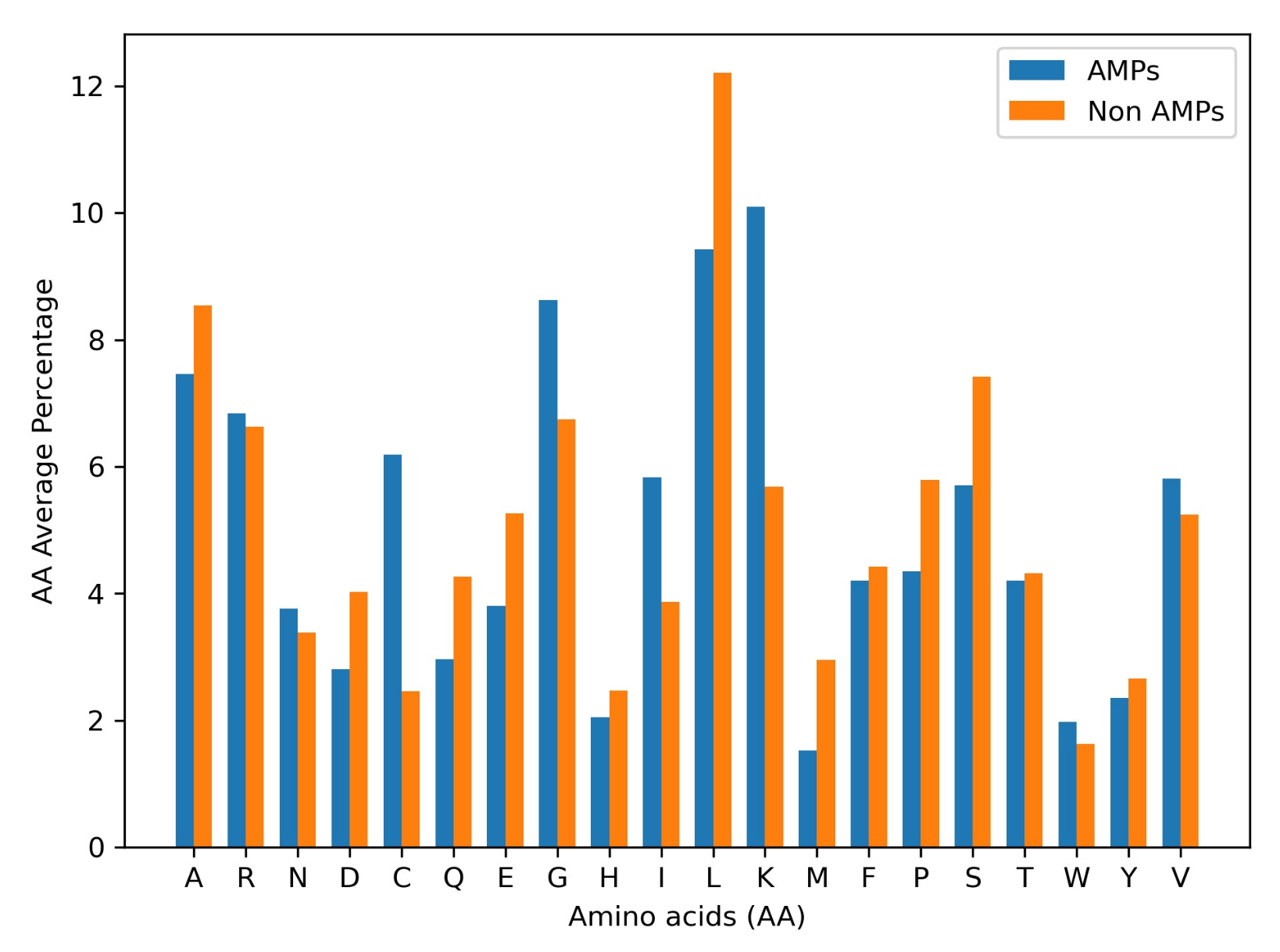
2.1. Database
- Peptides with synthetic modifications were deleted.
- Peptides with unknown amino acids (X) within their sequence were deleted.
- Peptides with pyrrolysine (O), Selenocysteine (U), -Alanine (Bal), 3-Naphthylalanine (Nal), and 2-Aminobutanoic acid (Abu) were deleted.
- Peptides with J (leucine or isoleucine) were maintained, considering both amino acids.
- Peptides with B (aspartic acid or Asparagine) were maintained, considering both amino acids.
- Peptides with Z (Glutamic acid or Glutamine) were maintained, considering both amino acids.
- Duplicated sequences were deleted while preserving all their associated activities.
2.1.1. AM Prediction
2.1.2. Fine AM Prediction
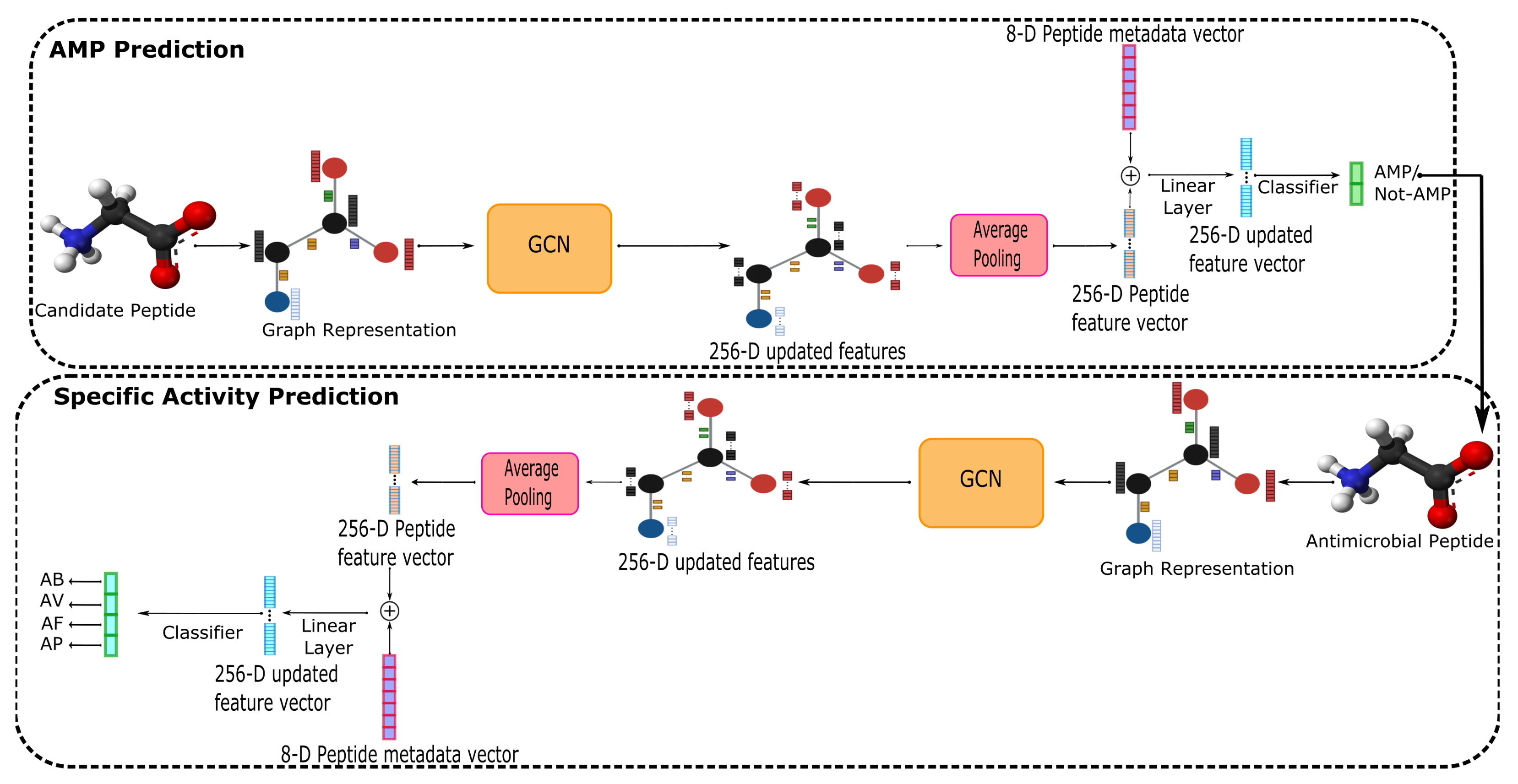
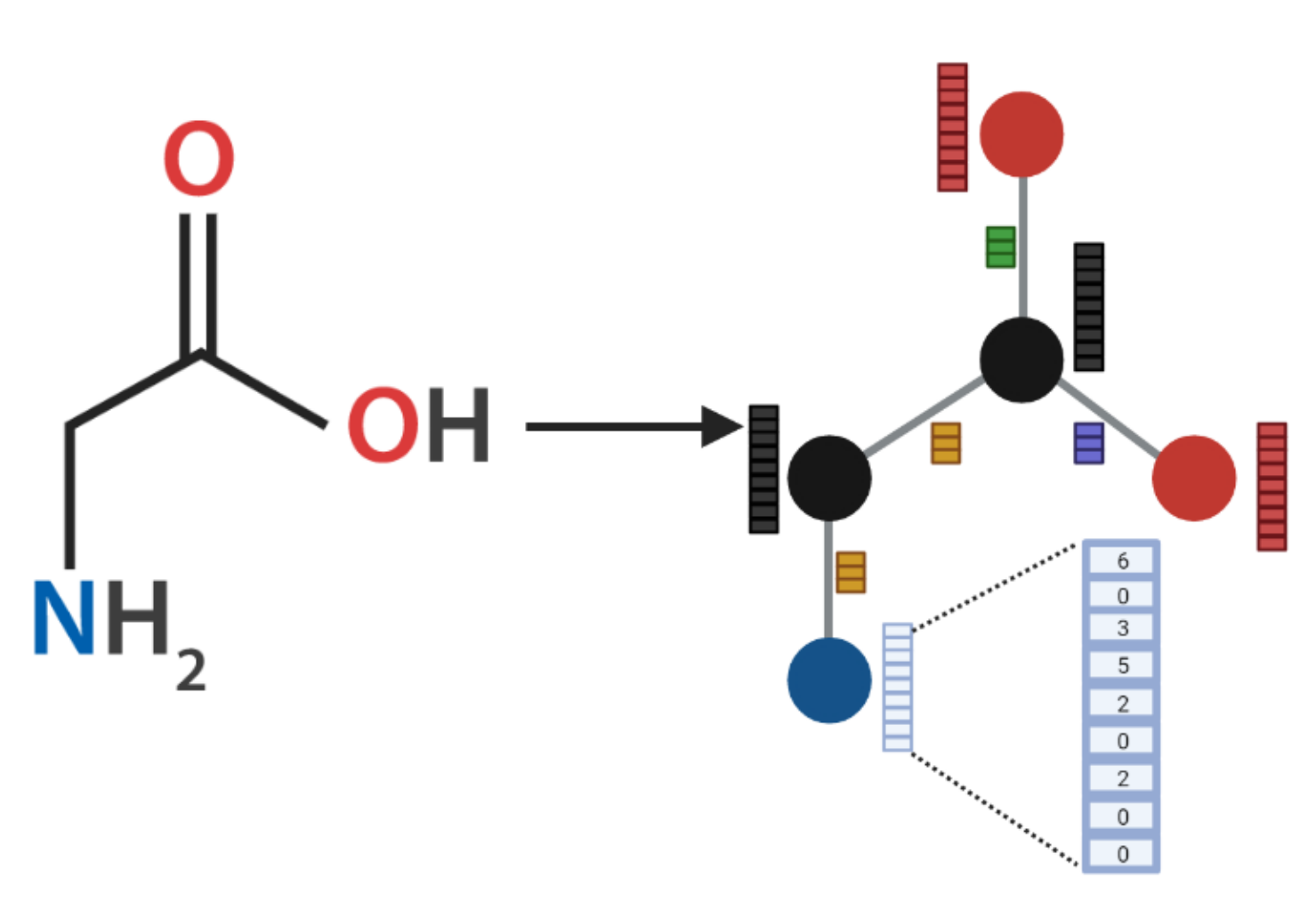
2.2. AMPs-Net
2.3. AMP Candidates
2.4. Molecular Dynamics Analysis
2.4.1. Non-Equilibrium Pulling (Flat-Bottom)
2.4.2. Non-Equilibrium Pulling (Umbrella SAMPLING)
2.4.3. Behavior Inside Membrane
2.5. In Vitro Validation
2.5.1. Antimicrobial Activity Validation
2.5.2. Synthesis of Low Molecular Weight Chitosan Nanoparticles (CNPs)
2.5.3. Functionalization of CNPs
2.5.4. Cell Penetrating Activity Validation
3. Results and Discussion
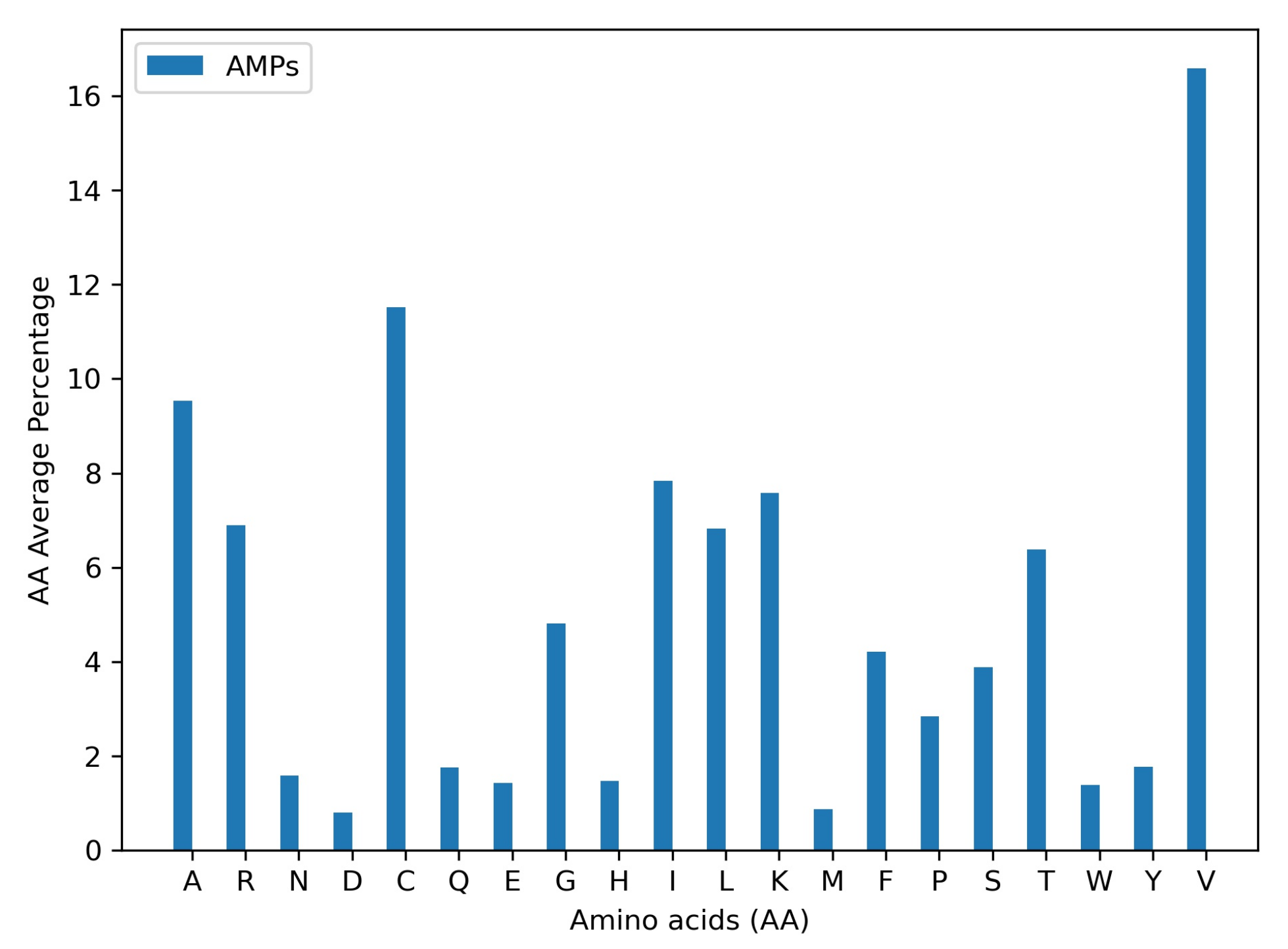
3.1. AMP Prediction
3.2. Candidates Selection
3.2.1. Monofunctional Peptides: AM Activity
3.2.2. Bifuctional Peptides: AM + CP Activity
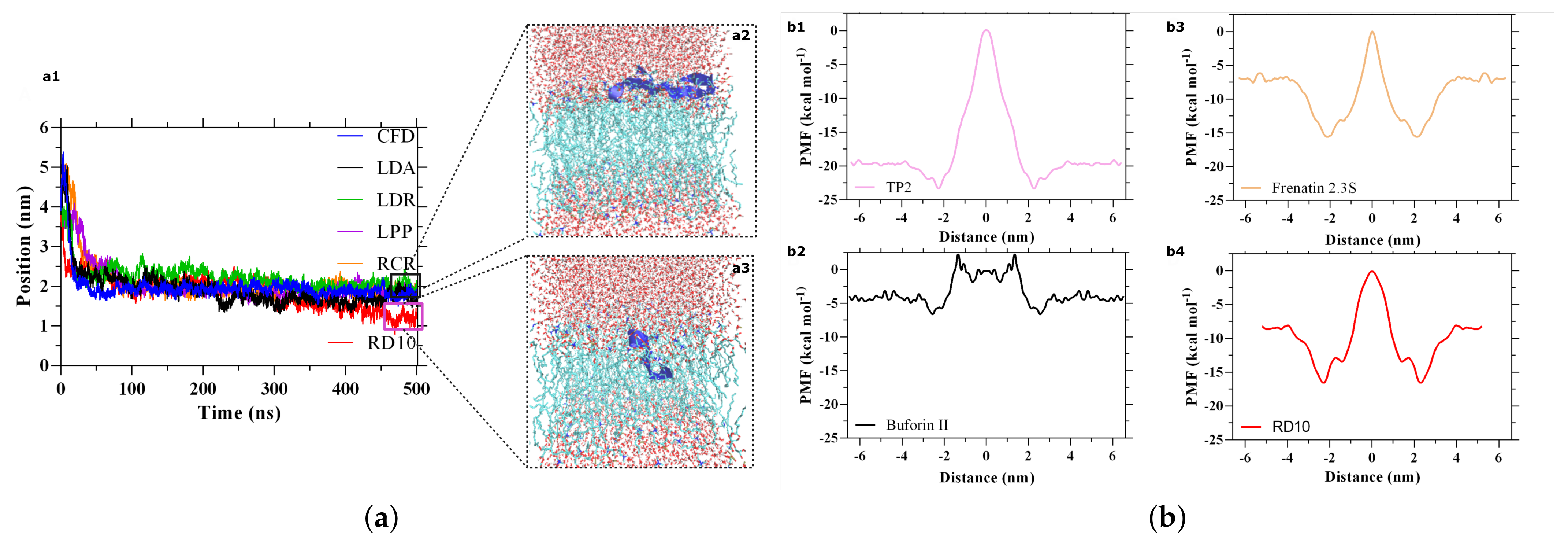
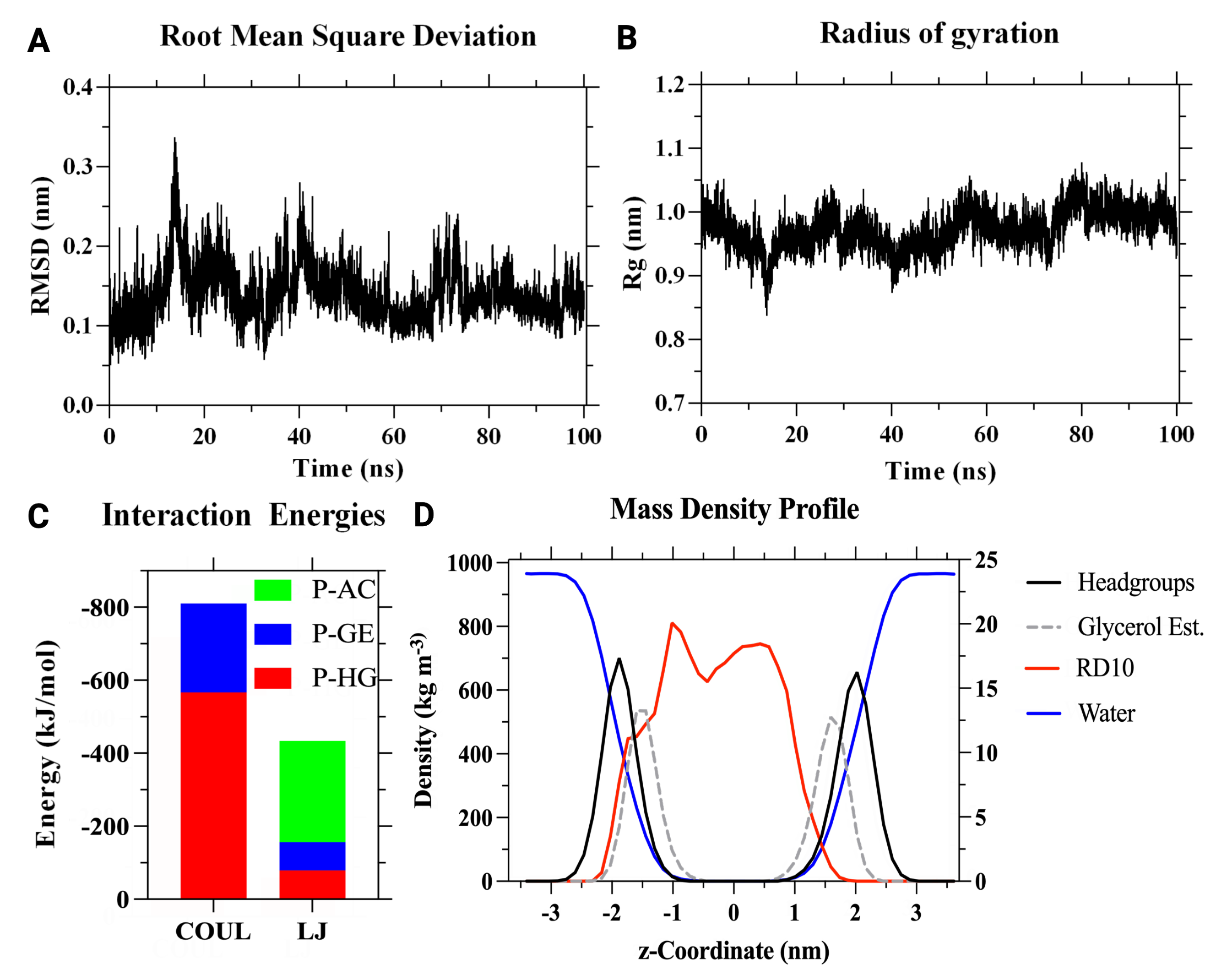
3.3. AM Validation
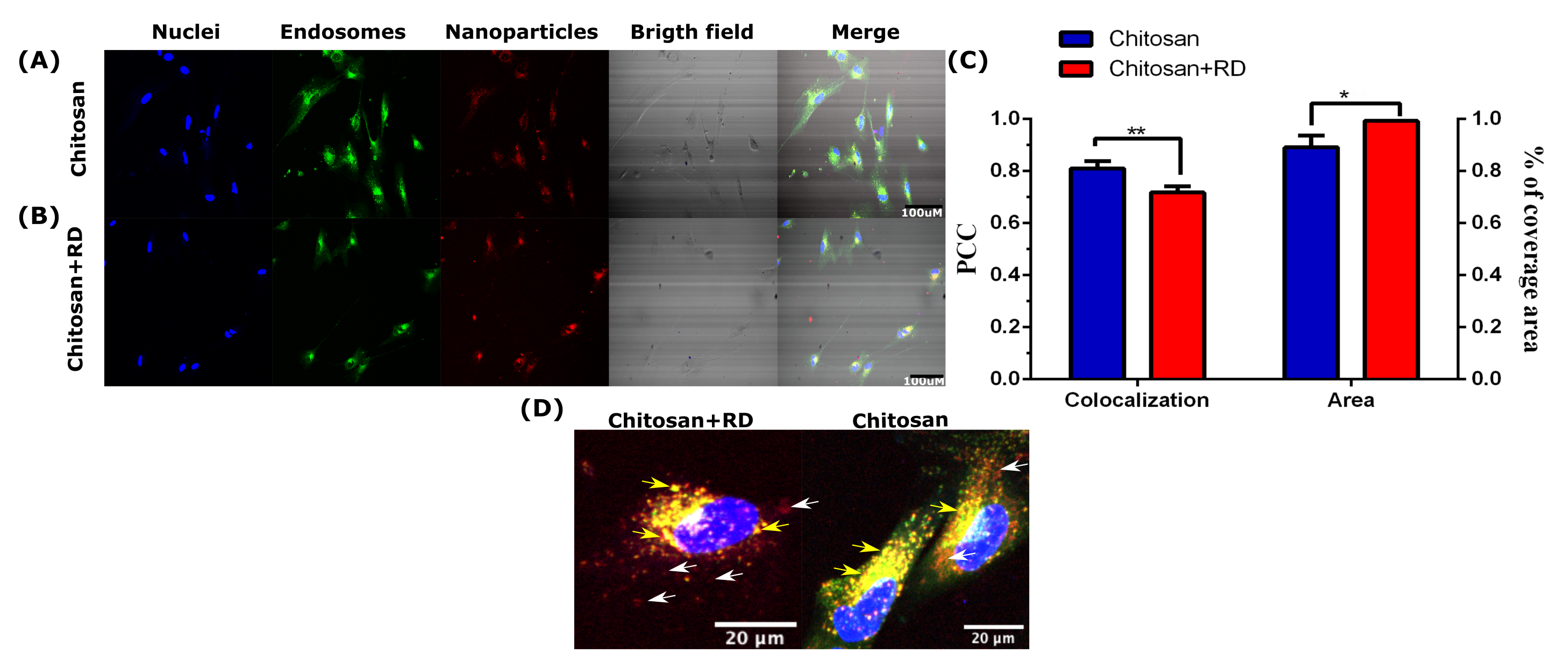
3.4. CP Validation
3.5. Clinical Applications
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaffiri, L.; Gardner, J.; Toledo-Pereyra, L.H. History of antibiotics. From salvarsan to cephalosporins. J. Invest. Surg. 2012, 25, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Naylor, N.R.; Atun, R.; Zhu, N.; Kulasabanathan, K.; Silva, S.; Chatterjee, A.; Knight, G.M.; Robotham, J.V. Estimating the burden of antimicrobial resistance: A systematic literature review. Antimicrob. Resist. Infect. Control 2018, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Stokowski, L.A. Antimicrobial Resistance: A Primer. 2010. Available online: https://www.medscape.com/viewarticle/729196_2 (accessed on 15 November 2021).
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. About Antibiotic Resistance. 2019. Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 15 November 2021).
- World Health Organization. Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- El-Mahallawy, H.A.; Hassan, S.S.; El-Wakil, M.; Moneer, M.M. Bacteremia due to ESKAPE pathogens: An emerging problem in cancer patients. J. Egypt. Natl. Cancer Inst. 2016, 28, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B. The SMART model: Soft Membranes Adapt and Respond, also Transiently, in the presence of antimicrobial peptides. J. Pept. Sci. 2014, 21, 346–355. [Google Scholar] [CrossRef] [Green Version]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Diener, C.; Garza Ramos Martínez, G.; Moreno Blas, D.; Castillo González, D.A.; Corzo, G.; Castro-Obregon, S.; Del Rio, G. Effective Design of Multifunctional Peptides by Combining Compatible Functions. PLoS Comput. Biol. 2016, 12, e1004786. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Boschin, G.; Arnoldi, A. Multifunctional peptides for the prevention of cardiovascular disease: A new concept in the area of bioactive food-derived peptides. J. Funct. Foods 2019, 55, 135–145. [Google Scholar] [CrossRef]
- Martin-Gómez, H.; Oliver-Cervelló, L.; Buxadera-Palomero, J.; Ginebra, M.P.; Mas-Moruno, C. Chemically Diverse Multifunctional Peptide Platforms with Antimicrobial and Cell Adhesive Properties. ChemBioChem 2020, 22, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Oliver-Cervelló, L.; Martin-Gómez, H.; Mas-Moruno, C. New trends in the development of multifunctional peptides to functionalize biomaterials. J. Pept. Sci. 2021, 28, e3335. [Google Scholar] [CrossRef]
- Li, M.; Guan, Y.; Zhao, A.; Ren, J.; Qu, X. Using Multifunctional Peptide Conjugated Au Nanorods for Monitoring β-amyloid Aggregation and Chemo-Photothermal Treatment of Alzheimer’s Disease. Theranostics 2017, 7, 2996–3006. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Li, Q.; Guo, J.; Ren, X.; Feng, Y.; Shi, C.; Zhang, W. Multifunctional Gene Carriers with Enhanced Specific Penetration and Nucleus Accumulation to Promote Neovascularization of HUVECs in Vivo. ACS Appl. Mater. Interfaces 2017, 9, 35613–35627. [Google Scholar] [CrossRef]
- Cheng, Y.; Sun, C.; Liu, R.; Yang, J.; Dai, J.; Zhai, T.; Lou, X.; Xia, F. A Multifunctional Peptide-Conjugated AIEgen for Efficient and Sequential Targeted Gene Delivery into the Nucleus. Angew. Chem. 2019, 131, 5103–5107. [Google Scholar] [CrossRef]
- Li, K.; Liu, C.J.; Zhang, X.Z. Multifunctional peptides for tumor therapy. Adv. Drug Deliv. Rev. 2020, 160, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Kula, E.; Kocazorbaz, E.K.; Moulahoum, H.; Alpat, S.; Zihnioglu, F. Extraction and characterization of novel multifunctional peptides from Trachinus Draco (greater weever) myofibrillar proteins with ACE/DPP4 inhibitory, antioxidant, and metal chelating activities. J. Food Biochem. 2020, 44, e13179. [Google Scholar] [CrossRef]
- Paray, B.A.; Ahmad, A.; Khan, J.M.; Taufiq, F.; Pathan, A.; Malik, A.; Ahmed, M.Z. The role of the multifunctional antimicrobial peptide melittin in gene delivery. Drug Discov. Today 2021, 26, 1053–1059. [Google Scholar] [CrossRef]
- Boas, L.C.P.V.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef]
- Kalafatovic, D.; Mauša, G.; Todorovski, T.; Giralt, E. Algorithm-supported, mass and sequence diversity-oriented random peptide library design. J. Cheminformat. 2019, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Quan, Z.; Wang, Z.J.; Huang, H.; Zeng, X. A novel molecular representation with BiGRU neural networks for learning atom. Brief. Bioinform. 2019, 21, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 13 October 2021).
- Sébastien Giguère, F.L. Machine Learning Assisted Design of Highly Active Peptides for Drug Discovery. PLoS Comput. Biol. 2015, 11, e1004074. [Google Scholar]
- Lane, N.; Kahanda, I. DeepACPpred: A Novel Hybrid CNN-RNN Architecture for Predicting Anti-Cancer Peptides. In Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2020; pp. 60–69. [Google Scholar]
- Yi, H.C.; You, Z.H.; Zhou, X.; Cheng, L.; Li, X.; Jiang, T.H.; Chen, Z.H. ACP-DL: A Deep Learning Long Short-Term Memory Model to Predict Anticancer Peptides Using High-Efficiency Feature Representation. Mol. Ther. Nucleic Acids 2019, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Zeng, W.F.; Zhou, X.X.; Zhou, W.J.; Chi, H.; Zhan, J.; He, S.M. MS/MS Spectrum Prediction for Modified Peptides Using pDeep2 Trained by Transfer Learning. Anal. Chem. 2019, 91, 9724–9731. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [Green Version]
- Hamid, M.N.; Friedberg, I. Identifying antimicrobial peptides using word embedding with deep recurrent neural networks. Bioinformatics 2018, 35, 2009–2016. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef]
- Ruiz Puentes, P.; Henao, M.C.; Torres, C.E.; Gómez, S.C.; Gómez, L.A.; Burgos, J.C.; Arbeláez, P.; Osma, J.F.; Muñoz-Camargo, C.; Reyes, L.H.; et al. Design, Screening, and Testing of Non-Rational Peptide Libraries with Antimicrobial Activity: In Silico and Experimental Approaches. Antibiotics 2020, 9, 854. [Google Scholar] [CrossRef]
- Ruiz Puentes, P.; Rueda-Gensini, L.; Valderrama, N.; Hernández, I.; González, C.; Daza, L.; Muñoz-Camargo, C.; Cruz, J.C.; Arbeláez, P. Predicting target–ligand interactions with graph convolutional networks for interpretable pharmaceutical discovery. Sci. Rep. 2022, 12, 8434. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, S.; Agrawal, P.; Chaudhary, K.; Usmani, S.S.; Raghava, G.; Singh, S.; Gautam, A. CPPsite 2.0 Database of Cell-Penetrating Peptides; Indraprastha Institute of Information Technology: New Delhi, India, 2015. [Google Scholar]
- Waghu, F.H.; Idicula-Thomas, S. Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci. 2019, 29, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, P.; Singh, H.; Gautam, A.; Chaudhary, K.; Kumar, R.; Raghava, G.P.S. TumorHoPe: A Database of Tumor Homing Peptides. PLoS ONE 2012, 7, e35187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2015, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choo, K.; Tan, T.; Ranganathan, S. SPdb—A signal peptide database. BMC Bioinform. 2005, 6, 249. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, S.; Agrawal, P.; Chaudhary, K.; Usmani, S.S.; Raghava, G.; Singh, S.; Gautam, A. ParaPep—A Database of Anti-Parasitic Peptides; Indraprastha Institute of Information Technology: New Delhi, India, 2014. [Google Scholar]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [Green Version]
- Dorpe, S.V.; Bronselaer, A.; Nielandt, J.; Stalmans, S.; Wynendaele, E.; Audenaert, K.; Wiele, C.V.D.; Burvenich, C.; Peremans, K.; Hsuchou, H.; et al. Brainpeps: The blood–brain barrier peptide database. Brain Struct. Funct. 2011, 217, 687–718. [Google Scholar] [CrossRef]
- Wynendaele, E.; Bronselaer, A.; Nielandt, J.; D’Hondt, M.; Stalmans, S.; Bracke, N.; Verbeke, F.; Wiele, C.V.D.; Tré, G.D.; Spiegeleer, B.D. Quorumpeps database: Chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2012, 41, D655–D659. [Google Scholar] [CrossRef]
- Piotto, S.; Sessa, L.; Concilio, S.; Iannelli, P. YADAMP: Yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents 2012, 39, 346–351. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, H.; Lu, H.; Li, G.; Huang, Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS ONE 2013, 8, e66557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théolier, J.; Fliss, I.; Jean, J.; Hammami, R. MilkAMP: A comprehensive database of antimicrobial peptides of dairy origin. Dairy Sci. Technol. 2014, 94, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Novković, M.; Simunić, J.; Bojović, V.; Tossi, A.; Juretić, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Kumar, R.; Kumar, V.; Singh, S.; Raghava, G.P.S. AntiTbPdb: A knowledgebase of anti-tubercular peptides. Database 2018, 2018, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Data Analysis & Modeling Group at Hasselt University and Functional Genomics and Proteomics Unit at K.U. Leuven. PeptideDB: Bioactive Peptide Database; Leuven, Belgium, 2022; Available online: http://www.peptides.be/?p=contact (accessed on 13 June 2022).
- Wang, Y.; Wang, M.; Yin, S.; Jang, R.; Wang, J.; Xue, Z.; Xu, T. NeuroPep: A Comprehensive Resource of Neuropeptides. Database 2015, 2015, bav038. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2015, 44, D1119–D1126. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kizhakkedathu, J.; Straus, S. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Xiong, C.; Thabet, A.; Ghanem, B. DeeperGCN: All You Need to Train Deeper GCNs. arXiv 2020, arXiv:2006.07739. [Google Scholar]
- McDonal, D.B.; Potts, W.K. DNA microsatellites as genetic markers at several scales. In Avian Molecular Evolution and Systematics; Elsevier: Amsterdam, The Netherlands, 1997; pp. 29–49. [Google Scholar] [CrossRef]
- Henao, M.C.; Ocasion, C.; Ruiz Puentes, P.; González-Melo, C.; Quezada, V.; Cifuentes, J.; Yepes, A.; Burgos, J.C.; Cruz, J.C.; Reyes, L.H. Translocating Peptides of Biomedical Interest Obtained from the Spike (S) Glycoprotein of the SARS-CoV-2. Membranes 2022, 12, 600. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Berger, O.; Edholm, O.; Jähnig, F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997, 72, 2002–2013. [Google Scholar] [CrossRef] [Green Version]
- Lemkul, J. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Perez, J.; Cifuentes, J.; Cuellar, M.; Suarez-Arnedo, A.; Cruz, J.C.; Muñoz-Camargo, C. Cell-Penetrating And Antibacterial BUF-II Nanobioconjugates: Enhanced Potency Via Immobilization On Polyetheramine-Modified Magnetite Nanoparticles. Int. J. Nanomed. 2019, 14, 8483–8497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Melo, C.; Garcia-Brand, A.J.; Quezada, V.; Reyes, L.H.; Muñoz-Camargo, C.; Cruz, J.C. Highly Efficient Synthesis of Type B Gelatin and Low Molecular Weight Chitosan Nanoparticles: Potential Applications as Bioactive Molecule Carriers and Cell-Penetrating Agents. Polymers 2021, 13, 4078. [Google Scholar] [CrossRef]
- Lin, T.T.; Yang, L.Y.; Lu, I.H.; Cheng, W.C.; Hsu, Z.R.; Chen, S.H.; Lin, C.Y. AI4AMP: An Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning. mSystems 2021, 6, e00299-21. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides: Table 1. Nucleic Acids Res. 2015, 44, D1094–D1097. [Google Scholar] [CrossRef] [Green Version]
- Pinacho-Castellanos, S.A.; García-Jacas, C.R.; Gilson, M.K.; Brizuela, C.A. Alignment-Free Antimicrobial Peptide Predictors: Improving Performance by a Thorough Analysis of the Largest Available Data Set. J. Chem. Inf. Model. 2021, 61, 3141–3157. [Google Scholar] [CrossRef] [PubMed]
- Pinacho-Castellanos, S.A.; García-Jacas, C.R.; Gilson, M.K.; Brizuela, C.A. AMPDiscover; CICESE: Baja California, Mexico, 2021. [Google Scholar]
- Lawrence, T.J.; Carper, D.L.; Spangler, M.K.; Carrell, A.A.; Rush, T.A.; Minter, S.J.; Weston, D.J.; Labbé, J.L. amPEPpy 1.0: A portable and accurate antimicrobial peptide prediction tool. Bioinformatics 2020, 37, 2058–2060. [Google Scholar] [CrossRef] [PubMed]
- Perumal, P.; Pandey, V.P. Antimicrobial peptides: The role of hydrophobicity in the alpha helical structure. J. Pharm. Pharmacogn. Res. 2013, 1, 39–53. [Google Scholar]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R J. 2017, 7, 4–14. [Google Scholar] [CrossRef]
- Marc, T.; David, A.; Nogues, V.M.; Ester, B. Connecting Peptide Physicochemical and Antimicrobial Properties by a Rational Prediction Model. PLoS ONE 2011, 6, e16968. [Google Scholar] [CrossRef]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Somma, A.D.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef] [PubMed]
- Nam, B.H.; Moon, J.Y.; Park, E.H.; Kim, Y.O.; Kim, D.G.; Kong, H.J.; Kim, W.J.; Jee, Y.J.; An, C.M.; Park, N.G.; et al. Antimicrobial Activity of Peptides Derived from Olive Flounder Lipopolysaccharide Binding Protein/Bactericidal Permeability-Increasing Protein (LBP/BPI). Mar. Drugs 2014, 12, 5240–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.A.; Huttunen-Hennelly, H.E.K.; Friedman, C.R. Bioactivity and the First Transmission Electron Microscopy Immunogold Studies of Short De Novo-Designed Antimicrobial Peptides. Antimicrob. Agents Chemother. 2011, 55, 2137–2145. [Google Scholar] [CrossRef] [Green Version]
- Bartłomiej, D.; Marta, D. New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies. Int. J. Mol. Sci. 2014, 15, 14531–14545. [Google Scholar] [CrossRef] [Green Version]
- Li, R.F.; Lu, Z.F.; Sun, Y.N.; Chen, S.H.; Yi, Y.J.; Zhang, H.R.; Yang, S.Y.; Yu, G.H.; Huang, L.; Li, C.N. Molecular Design, Structural Analysis and Antifungal Activity of Derivatives of Peptide CGA-N46. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Camargo, C.; Mitran, E.; Groot, H. Busqueda de Peptidos Antimicrobianos Nuevos en Secreciones de Piel de Ranas; Universidad de los Andes: Bogota, Colombia, 2015. [Google Scholar]
- Mor, A.; Amiche, M.; Nicolas, P. Structure, synthesis, and activity of Dermaseptin b, a novel vertebrate defensive peptide from frog skin: Relationship with adenoregulin. Biochemistry 1994, 33, 6642–6650. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Mor, A.; Nicolas, P.; Shai, Y. Spectrum of Antimicrobial Activity and Assembly of Dermaseptin-b and Its Precursor Form in Phospholipid Membranes. Biochemistry 1994, 33, 10951–10960. [Google Scholar] [CrossRef]
- Charpentier, S.; Amiche, M.; Mester, J.; Vouille, V.; Le Caer, J.P.; Nicolas, P.; Delfour, A. Structure, Synthesis, and Molecular Cloning of Dermaseptins B, a Family of Skin Peptide Antibiotics. J. Biol. Chem. 1998, 273, 14690–14697. [Google Scholar] [CrossRef] [Green Version]
- Villa-Hernandez, O.; Hernandez-Orihuela, L.; Rodriguez, M.; Zamudio-Zuniga, F.; Castro-Franco, R.; Pando, V.; Batista, C. Novel Antimicrobial Peptides Isolated from Skin Secretions of the Mexican Frog Hyla eximia. Protein Pept. Lett. 2009, 16, 1371–1378. [Google Scholar] [CrossRef]
- Bartels, E.J.H.; Dekker, D.; Amiche, M. Dermaseptins, Multifunctional Antimicrobial Peptides: A Review of Their Pharmacology, Effectivity, Mechanism of Action, and Possible Future Directions. Front. Pharmacol. 2019, 10, 1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacková, Z.; Branny, P. Functional Study of the Putative Nucleotidase Encoded by spr1057 Gene in Streptococcus Pneumoniae, a Likely Homolog of Escherichia coli Protein YjjG; Univerzita Karlova, Přírodovědecká Fakulta: Staré Město, Czech Republic, 2010. [Google Scholar]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, R.E.; Rozek, A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 2002, 206, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Ulmschneider, J.P. Charged Antimicrobial Peptides Can Translocate across Membranes without Forming Channel-like Pores. Biophys. J. 2017, 113, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Camargo, C.; Salazar, V.; Barrero-Guevara, L.; Camargo, S.; Mosquera, A.; Groot, H.; Boix, E. Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus). Int. J. Mol. Sci. 2018, 19, 2170. [Google Scholar] [CrossRef] [Green Version]
- Cruz, J.; Mihailescu, M.; Wiedman, G.; Herman, K.; Searson, P.; Wimley, W.; Hristova, K. A Membrane-Translocating Peptide Penetrates into Bilayers without Significant Bilayer Perturbations. Biophys. J. 2013, 104, 2419–2428. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Takeshima, K.; Park, C.B.; Kim, S.C.; Matsuzaki, K. Interactions of the Novel Antimicrobial Peptide Buforin 2 with Lipid Bilayers: Proline as a Translocation Promoting Factor. Biochemistry 2000, 39, 8648–8654. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Y.; Liu, H. Molecular Dynamics Simulations of Peptides and Proteins with Amplified Collective Motions. Biophys. J. 2003, 84, 3583–3593. [Google Scholar] [CrossRef] [Green Version]
- Maiorov, V.N.; Crippen, G.M. Significance of Root-Mean-Square Deviation in Comparing Three-dimensional Structures of Globular Proteins. J. Mol. Biol. 1994, 235, 625–634. [Google Scholar] [CrossRef] [Green Version]
- Daura, X.; Mark, A.E.; van Gunsteren, W.F. Peptide folding simulations: No solvent required? Comput. Phys. Commun. 1999, 123, 97–102. [Google Scholar] [CrossRef]
- Arnittali, M.; Rissanou, A.N.; Harmandaris, V. Structure Of Biomolecules Through Molecular Dynamics Simulations. Procedia Comput. Sci. 2019, 156, 69–78. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Orozco, R.Q.; Rezende, S.B.; Rodrigues, G.; Oshiro, K.G.N.; Cândido, E.S.; Franco, O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020, 10, 3097. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R.E.W. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008–1012. [Google Scholar] [CrossRef]
- Rollema, H.S.; Kuipers, O.P.; Both, P.; de Vos, W.M.; Siezen, R.J. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 1995, 61, 2873–2878. [Google Scholar] [CrossRef] [Green Version]
- Loree, J. Bacteriostatic Antibiotics. 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK547678/ (accessed on 13 November 2021).
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef] [Green Version]
- Loffredo, M.R.; Savini, F.; Bobone, S.; Casciaro, B.; Franzyk, H.; Mangoni, M.L.; Stella, L. Inoculum effect of antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2021, 118, e2014364118. [Google Scholar] [CrossRef]
- Ghosh, S.; Pandit, G.; Debnath, S.; Chatterjee, S.; Satpati, P. Effect of monovalent salt concentration and peptide secondary structure in peptide-micelle binding. RSC Adv. 2021, 11, 36836–36849. [Google Scholar] [CrossRef]
- Kandasamy, S.K.; Larson, R.G. Effect of salt on the interactions of antimicrobial peptides with zwitterionic lipid bilayers. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Rueda-Gensini, L.; Cifuentes, J.; Castellanos, M.C.; Ruiz Puentes, P.; Serna, J.A.; Muñoz-Camargo, C.; Cruz, J.C. Tailoring Iron Oxide Nanoparticles for Efficient Cellular Internalization and Endosomal Escape. Nanomaterials 2020, 10, 1816. [Google Scholar] [CrossRef] [PubMed]
- Pankey, G.A.; Sabath, L.D. Clinical Relevance of Bacteriostatic versus Bactericidal Mechanisms of Action in the Treatment of Gram-Positive Bacterial Infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [Green Version]
- Wald-Dickler, N.; Holtom, P.; Spellberg, B. Busting the Myth of “Static vs Cidal”: A Systemic Literature Review. Clin. Infect. Dis. 2017, 66, 1470–1474. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, J.; Zhao, M.; Tang, S.; Cheng, X.; Zhang, W.; Li, W.; Liu, X.; Peng, H.; Wang, Q. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale 2021, 13, 10748–10764. [Google Scholar] [CrossRef]
- Fang, Y.P.; Wu, P.C.; Huang, Y.B.; Tzeng, C.C.; Chen, Y.L.; Hung, Y.H.; Tsai, M.J.; Wu, P.C. Modification of polyethylene glycol onto solid lipid nanoparticles encapsulating a novel chemotherapeutic agent (PK-L4) to enhance solubility for injection delivery. Int. J. Nanomed. 2012, 7, 4995. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Abetxuko, A.; Sánchez-deAlcázar, D.; Muñumer, P.; Beloqui, A. Tunable Polymeric Scaffolds for Enzyme Immobilization. Front. Bioeng. Biotechnol. 2020, 8, 830. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, T.; Chetia, M.; Chatterjee, S. Antimicrobial Peptides and Proteins: From Nature’s Reservoir to the Laboratory and Beyond. Front. Chem. 2021, 9, 432. [Google Scholar] [CrossRef]
- Yang, Z.; He, S.; Wu, H.; Yin, T.; Wang, L.; Shan, A. Nanostructured Antimicrobial Peptides: Crucial Steps of Overcoming the Bottleneck for Clinics. Front. Microbiol. 2021, 12, 710199. [Google Scholar] [CrossRef]
| Database Name | Number of Peptides |
|---|---|
| BIOPEP-UWM Database [37] | 3634 |
| CPPsite 2.0 [38] | 1155 |
| CAMP [39] | 4519 |
| TumorHoPe [40] | 787 |
| APD3 [41] | 3072 |
| SPdb [42] | 2512 |
| ParaPep [43] | 194 |
| CancerPPD [44] | 556 |
| BrainPreps [45] | 92 |
| Quorumpeps [46] | 257 |
| YADAMP [47] | 2525 |
| LAMP2 [48] | 5454 |
| Milkampdb [49] | 260 |
| DADP [50] | 2557 |
| AntiTbPdb [51] | 271 |
| PeptideDB [52] | 1903 |
| NeuroPrep [53] | 3875 |
| SATPdb [54] | 9664 |
| Other peptides | 1475 |
| Total | 44,762 |
| Biological Activity | Number of Peptides |
|---|---|
| Antimicrobial | 13,468 |
| Neuropeptide | 3615 |
| Signal-Peptide | 2351 |
| Anuran-Defense | 1783 |
| Anticancer | 1602 |
| Cell-Penetrating | 1155 |
| ACE-Inhibitor | 934 |
| TumorHoming | 704 |
| Antioxidative | 637 |
| Peptidase-IV-Inhibitor | 420 |
| Toxic | 256 |
| QuorumSensing-Peptide | 252 |
| Opioid | 136 |
| BBB-Peptide | 88 |
| Immunomodulating | 71 |
| Peptidase-III-Inhibitor | 66 |
| Haemolytic | 63 |
| Antithrombotic | 58 |
| Antiamnestic | 52 |
| CaMKII-Inhibitor | 50 |
| Insecticidal | 49 |
| Alpha-glucosidase-Inhibitor | 34 |
| Renin-Inhibitor | 19 |
| Atom Features | |
|---|---|
| Atomic Number | 1, 2, …, 119 |
| Chirality | Unspecified, Tetrahedral clockwise, Tetrahedral anti-clockwise, Other |
| Degree | 0, 1, …, 10 |
| Formal Charge | −5, −4, …, 4, 5 |
| Number of Hydrogens | 0, 1, …, 8 |
| Number of radical e | 0, 1, …, 4 |
| Hybridization | Sp, Sp, Sp, Spd, Spd |
| Aromaticity | 0, 1 |
| Ring membership | 0, 1 |
| Bond Features | |
| Type | Single, Double, Triple, Aromatic |
| Stereochemistry | None, Z, E, CIS, TRANS, Any |
| Conjugation | 0, 1 |
| Method | AP | ACC |
|---|---|---|
| AMPScanner [32] | 82.1 | 65.58 |
| AI4AMPs [64] | 76.74 | 67.64 |
| CAMP [65] | - | 67.82 |
| AMPDiscover [66,67] | - | 71.63 |
| AMPlify [30] | 86.96 | 75.07 |
| AMPs-Net (Ours) | 95.76 | 89.81 |
| Non-deep learning SOTA | ||
| AMPEPpy (RF) [68] | 97.37 | 90.33 |
| Parameter | Test AP |
|---|---|
| GCN layers | |
| 10 Layers | 94.48 |
| 15 Layers | 94.32 |
| 20 Layers | 95.04 |
| 25 Layers | 94.86 |
| 30 Layers | 94.6 |
| Features Hidden Size | |
| HS 32 | 91.52 |
| HS 64 | 94.12 |
| HS 128 | 94.67 |
| HS 256 | 95.04 |
| Aggregation Function MLPs | |
| 2 MLP | 94.72 |
| 3 MLP | 95.04 |
| 4 MLP | 95.09 |
| Metadata concatenation | |
| 8 Features | 95.76 |
| Antimicrobial Activity | AP |
|---|---|
| Antibacterial | 90.57 |
| Antiviral | 84.54 |
| Antifungal | 50.93 |
| Antiparasitic | 24.73 |
| Overall evaluation | NAP |
| Multiclass | 71.36 |
| Sequence | Size | Net Charge | Boman Index | Hydrophobic Ratio | Hydrophobic Moment | Aliphatic Index | Instability Index | Isoelectric Point |
|---|---|---|---|---|---|---|---|---|
| VFVVVTLLKKVKLLC (VC15) | 15 | 2.834 | −1.661 | 0.733 | 0.257 | 200.666 | −15.226 | 10.425 |
| KLKKVTGKKMSKCMKCKIYVCS (KS22) | 22 | 7.521 | 1.205 | 0.409 | 0.244 | 61.818 | 32.141 | 10.527 |
| RTLFVCRVGD (RD10) | 10 | 0.836 | 2.293 | 0.5 | 0.195 | 97.0 | 0.509 | 8.759 |
| FTFYLPLFVCRRNPRPRRVSCRE (FE23) | 23 | 4.68 | 3.463 | 0.391 | 0.240 | 59.103 | 107.39 | 11.428 |
| Sequence | NaPHO | NaCl | ||
|---|---|---|---|---|
| MIC (M) | MIC (M) | |||
| E. coli | S. aureus | E. coli | S. aureus | |
| VFVVVTLLKKVKLLC (VC15) | >160 | >160 | - | - |
| KLKKVTGKKMSKCMKCKIYVCS (KS22) | 250 | 250 | >250 | >250 |
| RTLFVCRVGD (RD10) | >250 | >250 | >250 | >250 |
| FTFYLPLFVCRRNPRPRRVSCRE (FE23) | 7.8 | 15.62 | >250 | >250 |
| Sequence | Concentration (M) | |
|---|---|---|
| E. coli | S. aureus | |
| VFVVVTLLKKVKLLC (VC15) | 80 | 80 |
| KLKKVTGKKMSKCMKCKIYVCS (KS22) | 7.8 | 7.8 |
| RTLFVCRVGD (RD10) | 0.48 | 0.48 |
| FTFYLPLFVCRRNPRPRRVSCRE (FE23) | 3.9 | 3.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz Puentes, P.; Henao, M.C.; Cifuentes, J.; Muñoz-Camargo, C.; Reyes, L.H.; Cruz, J.C.; Arbeláez, P. Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence. Membranes 2022, 12, 708. https://doi.org/10.3390/membranes12070708
Ruiz Puentes P, Henao MC, Cifuentes J, Muñoz-Camargo C, Reyes LH, Cruz JC, Arbeláez P. Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence. Membranes. 2022; 12(7):708. https://doi.org/10.3390/membranes12070708
Chicago/Turabian StyleRuiz Puentes, Paola, Maria C. Henao, Javier Cifuentes, Carolina Muñoz-Camargo, Luis H. Reyes, Juan C. Cruz, and Pablo Arbeláez. 2022. "Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence" Membranes 12, no. 7: 708. https://doi.org/10.3390/membranes12070708
APA StyleRuiz Puentes, P., Henao, M. C., Cifuentes, J., Muñoz-Camargo, C., Reyes, L. H., Cruz, J. C., & Arbeláez, P. (2022). Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence. Membranes, 12(7), 708. https://doi.org/10.3390/membranes12070708










