Odor Discrimination by Lipid Membranes
Abstract
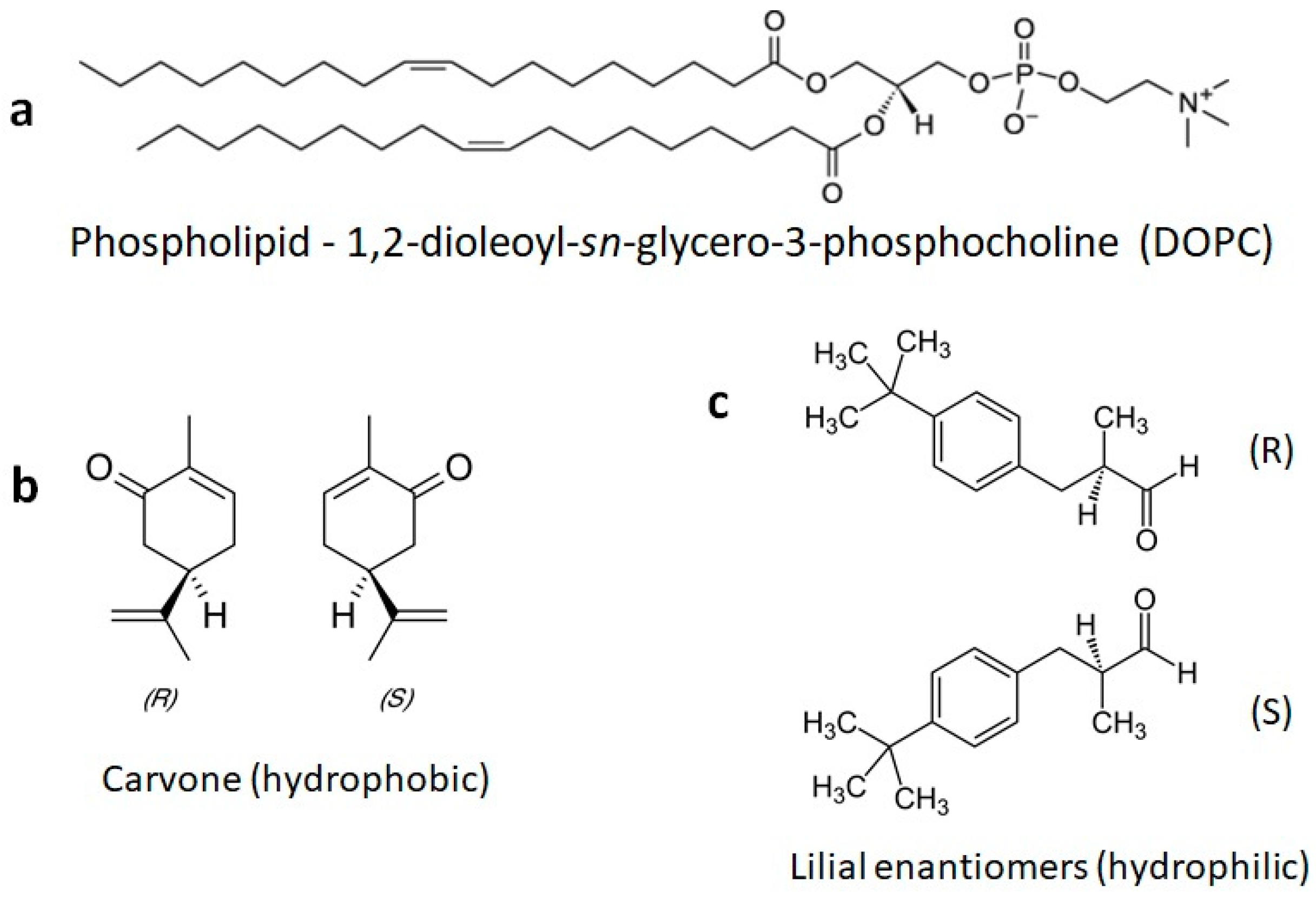
1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Firestein, S.; Darrow, B.; Shepherd, G.M. Activation of the sensory current in salamander olfactory receptor neurons depends on a G protein-mediated cAMP second messenger system. Neuron 1991, 6, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Reed, R.R. Golf: An olfactory neuron specific-G protein involved in odorant signal transduction. Science 1989, 244, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Masters, S.B.; Bourne, H.R.; Reed, R.R. Biochemical characterization of three stimulatory GTP-binding proteins. The large and small forms of Gs and the olfactory-specific G-protein, Golf. J. Biol. Chem. 1990, 265, 2671–2676. [Google Scholar] [CrossRef]
- Bruch, R.C. Signal transducing GTP-binding proteins in olfaction. Comp. Biochem. Physiol. A-Physiol. 1990, 95, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Dewan, A.; Cichy, A.; Zhang, J.; Miguel, K.; Feinstein, P.; Rinberg, D.; Bozza, T. Single olfactory receptors set odor detection thresholds. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Geithe, C.; Protze, J.; Kreuchwig, F.; Krause, G.; Krautwurst, D. Structural determinants of a conserved enantiomer-selective carvone binding pocket in the human odorant receptor OR1A1. Cell. Mol. Life Sci. 2017, 74, 4209–4229. [Google Scholar] [CrossRef]
- Minic, J.; Persuy, M.-A.; Godel, E.; Aioun, J.; Connerton, I.; Salesse, R.; Pajot-Augy, E. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2004, 272, 524–537. [Google Scholar] [CrossRef]
- Sanmartí-Espinal, M.; Iavicoli, P.; Calò, A.; Taulés, M.; Galve, R.; Marco, M.P.; Samitier, J. Quantification of interacting cognate odorants with olfactory receptors in nanovesicles. Sci. Rep. 2017, 7, 17483. [Google Scholar] [CrossRef]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J. Odor coding by a mammalian receptor repertoire. Neurosci. Res. 2009, 65, S76. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, Y.; Matsunami, H.; Zhuang, H. Live-cell Measurement of Odorant Receptor Activation Using a Real-time cAMP Assay. J. Vis. Exp. 2017, 128, e55831. [Google Scholar] [CrossRef] [PubMed]
- Glatz, R.; Bailey-Hill, K. Mimicking nature’s noses: From receptor deorphaning to olfactory biosensing. Prog. Neurobiol. 2011, 93, 270–296. [Google Scholar] [CrossRef] [PubMed]
- Grigorieff, N.; Ceska, T.; Downing, K.; Baldwin, J.; Henderson, R. Electron-crystallographic Refinement of the Structure of Bacteriorhodopsin. J. Mol. Biol. 1996, 259, 393–421. [Google Scholar] [CrossRef]
- Butterwick, J.A.; Del Mármol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Briand, L.; A De March, C.; Matsunami, H.; Yamashita, A.; Meyerhof, W.; Weyand, S. Structure–Function Relationships of Olfactory and Taste Receptors. Chem. Senses 2018, 43, 81–87. [Google Scholar] [CrossRef]
- Man, O.; Gilad, Y.; Lancet, D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004, 13, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wu, B. Structural studies of G protein-coupled receptors. IUBMB Life 2016, 68, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Floriano, W.B.; Vaidehi, N.; Goddard, W.A.; Singer, M.S.; Shepherd, G.M. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc. Natl. Acad. Sci. USA 2000, 97, 10712–10716. [Google Scholar] [CrossRef]
- de March, C.A.; Yu, Y.; Ni, M.J.; Adipietro, K.A.; Matsunami, H.; Ma, M.; Golebiowski, J. Conserved Residues Control Activation of Mammalian G Protein-Coupled Odorant Receptors. J. Am. Chem. Soc. 2015, 137, 8611–8616. [Google Scholar] [CrossRef]
- Kepchia, D.; Sherman, B.; Haddad, R.; Luetje, C.W. Mammalian odorant receptor tuning breadth persists across distinct odorant panels. PLoS ONE 2017, 12, e0185329. [Google Scholar] [CrossRef]
- Boesveldt, S.; Postma, E.M.; Boak, D.; Welge-Luessen, A.; Schopf, V.; Mainland, J.D.; Martens, J.; Ngai, J.; Duffy, V.B. Anosmia-A Clinical Review. Chem. Senses 2017, 42, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Silva Teixeira, C.S.; Cerqueira, N.M.; Silva Ferreira, A.C. Unravelling the Olfactory Sense: From the Gene to Odor Perception. Chem. Senses 2016, 41, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, W.; Frazer, J.; Unett, D. Functional assays for screening GPCR targets. Curr. Opin. Biotechnol. 2005, 16, 655–665. [Google Scholar] [CrossRef]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef]
- Lee, A.G. How lipids affect the activities of integral membrane proteins. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2004, 1666, 62–87. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2009, 327, 46–50. [Google Scholar] [CrossRef]
- Sukharev, S.I.; Blount, P.; Martinac, B.; Blattner, F.R.; Kung, C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 1994, 368, 265–268. [Google Scholar] [CrossRef]
- Phillips, R.; Ursell, T.; Wiggins, P.; Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature 2009, 459, 379–385. [Google Scholar] [CrossRef]
- Mohole, M.; Sengupta, D.; Chattopadhyay, A. Synergistic and Competitive Lipid Interactions in the Serotonin1A Receptor Microenvironment. ACS Chem. Neurosci. 2022, 13, 3403–3415. [Google Scholar] [CrossRef]
- Sarkar, P.; Chattopadhyay, A. Cholesterol in GPCR Structures: Prevalence and Relevance. J. Membr. Biol. 2021, 255, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Oates, J.; Watts, A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 2011, 21, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Zhang, X.; Kong, W.; Yang, X.-L.; Molnár, G.; Vondráková, Z.; Filepová, R.; Petrášek, J.; Friml, J.; Xue, H.-W. The lipid code-dependent phosphoswitch PDK1–D6PK activates PIN-mediated auxin efflux in Arabidopsis. Nat. Plants 2020, 6, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Kahana, A.; Maslov, S.; Lancet, D. Dynamic lipid aptamers: Non-polymeric chemical path to early life. Chem. Soc. Rev. 2021, 50, 11741–11746. [Google Scholar] [CrossRef]
- Bell, T.N.; Feng, K.; Calvin, G.; Van Winkle, D.H.; Lenhert, S. Organic Composomes as Supramolecular Aptamers. ACS Omega 2020, 5, 27393–27400. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Kurihara, K. Effect of Odorants on Lipid Monolayers from Bovine Olfactory Epithelium. Nature 1972, 236, 402–404. [Google Scholar] [CrossRef] [PubMed]
- Fujita, R.; Yotsumoto, M.; Yamaguchi, Y.; Matsuo, M.; Fukuhara, K.; Takahashi, O.; Nakanishi, S.; Denda, M.; Nakata, S. Masking of a malodorous substance on 1,2-dioleoyl-sn-glycero-3-phosphocholine molecular layer. Colloids Surf. A-Physicochem. Eng. Asp. 2022, 634, 128045. [Google Scholar] [CrossRef]
- Wyszynski, B.; Somboon, P.; Nakamoto, T. Mixed self-assembled lipopolymers with spacer lipids enhancing sensitivity of lipid-derivative QCMs for odor sensors. Sens. Actuators B Chem. 2008, 134, 72–78. [Google Scholar] [CrossRef]
- Kashiwayanagi, M.; Sasaki, K.; Lida, A.; Saito, H.; Kurihara, K. Concentration and Membrane Fluidity Dependence of Odor Discrimination in the Turtle Olfactory System. Chem. Senses 1997, 22, 553–563. [Google Scholar] [CrossRef]
- Martín, F.; Riveron, J.; Alcorta, E. Environmental temperature modulates olfactory reception in Drosophila melanogaster. J. Insect Physiol. 2011, 57, 1631–1642. [Google Scholar] [CrossRef]
- Castro, T.; Silva, C.; Matamá, T.; Cavaco-Paulo, A. The Structural Properties of Odorants Modulate Their Association to Human Odorant Binding Protein. Biomolecules 2021, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Magna, G.; Stefanelli, M.; Pomarico, G.; Naitana, M.L.; Monti, D.; Di Natale, C.; Paolesse, R. Chiral Recognition with Broad Selective Sensor Arrays. Chemosensors 2022, 10, 308. [Google Scholar] [CrossRef]
- Chan, Y.-H.M.; Boxer, S.G. Model membrane systems and their applications. Curr. Opin. Chem. Biol. 2007, 11, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Meker, S.; Halevi, O.; Chin, H.; Sut, T.N.; Jackman, J.A.; Tan, E.-L.; Potroz, M.G.; Cho, N.-J. Inkjet-Printed Phospholipid Bilayers on Titanium Oxide Surfaces: Towards Functional Membrane Biointerfaces. Membranes 2022, 12, 361. [Google Scholar] [CrossRef]
- Pašalić, L.; Pem, B.; Bakarić, D. Lamellarity-Driven Differences in Surface Structural Features of DPPS Lipids: Spectroscopic, Calorimetric and Computational Study. Membranes 2023, 13, 83. [Google Scholar] [CrossRef]
- Loshkareva, A.S.; Popova, M.M.; Shilova, L.A.; Fedorova, N.V.; Timofeeva, T.A.; Galimzyanov, T.R.; Kuzmin, P.I.; Knyazev, D.G.; Batishchev, O.V. Influenza A Virus M1 Protein Non-Specifically Deforms Charged Lipid Membranes and Specifically Interacts with the Raft Boundary. Membranes 2023, 13, 76. [Google Scholar] [CrossRef]
- Babayco, C.B.; Turgut, S.; Smith, A.M.; Sanii, B.; Land, D.; Parikh, A.N. A comparison of lateral diffusion in supported lipid monolayers and bilayers. Soft Matter 2010, 6, 5877–5881. [Google Scholar] [CrossRef]
- Tero, R. Substrate Effects on the Formation Process, Structure and Physicochemical Properties of Supported Lipid Bilayers. Materials 2012, 5, 2658–2680. [Google Scholar] [CrossRef]
- Kusi-Appiah, A.E.; Lowry, T.W.; Darrow, E.M.; Wilson, K.A.; Chadwick, B.P.; Davidson, M.W.; Lenhert, S. Quantitative dose–response curves from subcellular lipid multilayer microarrays. Lab A Chip 2015, 15, 3397–3404. [Google Scholar] [CrossRef]
- Kusi-Appiah, A.E.; Vafai, N.; Cranfill, P.J.; Davidson, M.W.; Lenhert, S. Lipid multilayer microarrays for in vitro liposomal drug delivery and screening. Biomaterials 2012, 33, 4187–4194. [Google Scholar] [CrossRef]
- Kusi-Appiah, A.E.; Mastronardi, M.L.; Qian, C.; Chen, K.K.; Ghazanfari, L.; Prommapan, P.; Kübel, C.; Ozin, G.A.; Lenhert, S. Enhanced cellular uptake of size-separated lipophilic silicon nanoparticles. Sci. Rep. 2017, 7, srep43731. [Google Scholar] [CrossRef] [PubMed]
- Lenhert, S.; Brinkmann, F.; Laue, T.; Walheim, S.; Vannahme, C.; Klinkhammer, S.; Xu, M.; Sekula, S.; Mappes, T.; Schimmel, T.; et al. Lipid multilayer gratings. Nat. Nanotechnol. 2010, 5, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Lenhert, S.; Sun, P.; Wang, Y.; Fuchs, H.; Mirkin, C.A. Massively Parallel Dip-Pen Nanolithography of Heterogeneous Supported Phospholipid Multilayer Patterns. Small 2006, 3, 71–75. [Google Scholar] [CrossRef]
- Lowry, T.W.; Prommapan, P.; Rainer, Q.; Van Winkle, D.; Lenhert, S. Lipid Multilayer Grating Arrays Integrated by Nanointaglio for Vapor Sensing by an Optical Nose. Sensors 2015, 15, 20863–20872. [Google Scholar] [CrossRef]
- Lowry, T.W.; Hariri, H.; Prommapan, P.; Kusi-Appiah, A.; Vafai, N.; Bienkiewicz, E.A.; Van Winkle, D.H.; Stagg, S.M.; Lenhert, S. Quantification of Protein-Induced Membrane Remodeling Kinetics In Vitro with Lipid Multilayer Gratings. Small 2016, 12, 506–515. [Google Scholar] [CrossRef]
- Prommapan, P.; Brljak, N.; Lowry, T.; Van Winkle, D.; Lenhert, S. Aptamer Functionalized Lipid Multilayer Gratings for Label-Free Analyte Detection. Nanomaterials 2020, 10, 2433. [Google Scholar] [CrossRef] [PubMed]
- Nafday, A.; Lenhert, O.S. High-throughput optical quality control of lipid multilayers fabricated by dip-pen nanolithography. Nanotechnology 2011, 22, 225301. [Google Scholar] [CrossRef] [PubMed]
- Lowry, T.W.; Kusi-Appiah, A.; Guan, J.; Van Winkle, D.H.; Davidson, M.W.; Lenhert, S. Materials integration by nanointaglio. Adv. Mater. Interfaces 2014, 1, 1300127. [Google Scholar] [CrossRef]
- Sekula, S.; Fuchs, J.; Weg-Remers, S.; Nagel, P.; Schuppler, S.; Fragala, J.; Theilacker, N.; Franzreb, M.; Wingren, C.; Ellmark, P.; et al. Multiplexed lipid dip-pen nanolithography on subcellular scales for the templating of functional proteins and cell culture. Small 2008, 4, 1785–1793. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Kumar, R.; Takai, M.; Hirtz, M. Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer. Molecules 2020, 25, 2768. [Google Scholar] [CrossRef]
- Berganza, E.; Boltynjuk, E.; Mathew, G.; Vallejo, F.F.; Gröger, R.; Scherer, T.; Sekula-Neuner, S.; Hirtz, M. 3D Nanolithography by Means of Lipid Ink Spreading Inhibition. Small 2022, 2205590, Online ahead of print. [Google Scholar] [CrossRef]
- Kumar, R.; Urtizberea, A.; Ghosh, S.; Bog, U.; Rainer, Q.; Lenhert, S.; Fuchs, H.; Hirtz, M. Polymer Pen Lithography with Lipids for Large-Area Gradient Patterns. Langmuir 2017, 33, 8739–8748. [Google Scholar] [CrossRef] [PubMed]
- Vafai, N.; Lowry, T.W.; Wilson, K.A.; Davidson, M.W.; Lenhert, S. Evaporative edge lithography of a liposomal drug microarray for cell migration assays. Nanofabrication 2015, 2, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Nafday, O.A.; Lowry, T.W.; Lenhert, S. Multifunctional Lipid Multilayer Stamping. Small 2012, 8, 1021–1028. [Google Scholar] [CrossRef]
- Ghazanfari, L.; Lenhert, S. Screening of Lipid Composition for Scalable Fabrication of Solvent-Free Lipid Microarrays. Front. Mater. 2016, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Bergström, C.A.S.; Yazdanian, M. Lipophilicity in Drug Development: Too Much or Not Enough? AAPS J. 2016, 18, 1095–1100. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Lokugamage, M.; Sago, C.D.; Gan, Z.; Krupczak, B.; Dahlman, J.E. Constrained Nanoparticles Deliver siRNA and sgRNA to T Cells In Vivo without Targeting Ligands. Adv. Mater. 2019, 31, e1902251. [Google Scholar] [CrossRef]
- Bobo, D.; Robinson, K.J.; Islam, J.; Thurecht, K.J.; Corrie, S.R. Nanoparticle-Based Medicines: A Review of FDA-Approved Materials and Clinical Trials to Date. Pharm. Res. 2016, 33, 2373–2387. [Google Scholar] [CrossRef]
- Marchianò, V.; Matos, M.; López, M.; Weng, S.; Serrano-Pertierra, E.; Luque, S.; Blanco-López, M.C.; Gutiérrez, G. Nanovesicles as Vanillin Carriers for Antimicrobial Applications. Membranes 2023, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Selyutina, O.Y.; Mastova, A.V.; Polyakov, N.E. The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study. Membranes 2023, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Mutlu, A.S.; Gao, S.M.; Zhang, H.; Wang, M.C. Olfactory specificity regulates lipid metabolism through neuroendocrine signaling in Caenorhabditis elegans. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Tsuneki, H.; Sugiyama, M.; Ito, T.; Sato, K.; Matsuda, H.; Onishi, K.; Yubune, K.; Matsuoka, Y.; Nagai, S.; Yamagishi, T.; et al. Food odor perception promotes systemic lipid utilization. Nat. Metab. 2022, 4, 1514–1531. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lowry, T.W.; Kusi-Appiah, A.E.; Fadool, D.A.; Lenhert, S. Odor Discrimination by Lipid Membranes. Membranes 2023, 13, 151. https://doi.org/10.3390/membranes13020151
Lowry TW, Kusi-Appiah AE, Fadool DA, Lenhert S. Odor Discrimination by Lipid Membranes. Membranes. 2023; 13(2):151. https://doi.org/10.3390/membranes13020151
Chicago/Turabian StyleLowry, Troy W., Aubrey E. Kusi-Appiah, Debra Ann Fadool, and Steven Lenhert. 2023. "Odor Discrimination by Lipid Membranes" Membranes 13, no. 2: 151. https://doi.org/10.3390/membranes13020151
APA StyleLowry, T. W., Kusi-Appiah, A. E., Fadool, D. A., & Lenhert, S. (2023). Odor Discrimination by Lipid Membranes. Membranes, 13(2), 151. https://doi.org/10.3390/membranes13020151





