Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mutagenesis
2.2. Bacterial Growth and Protein Samples Preparation
2.3. Studies on the Properties of Reaction Centers
2.4. Polyacrylamide Gel Electrophoresis
2.5. Crystallization and X-ray Diffraction Analysis
3. Results
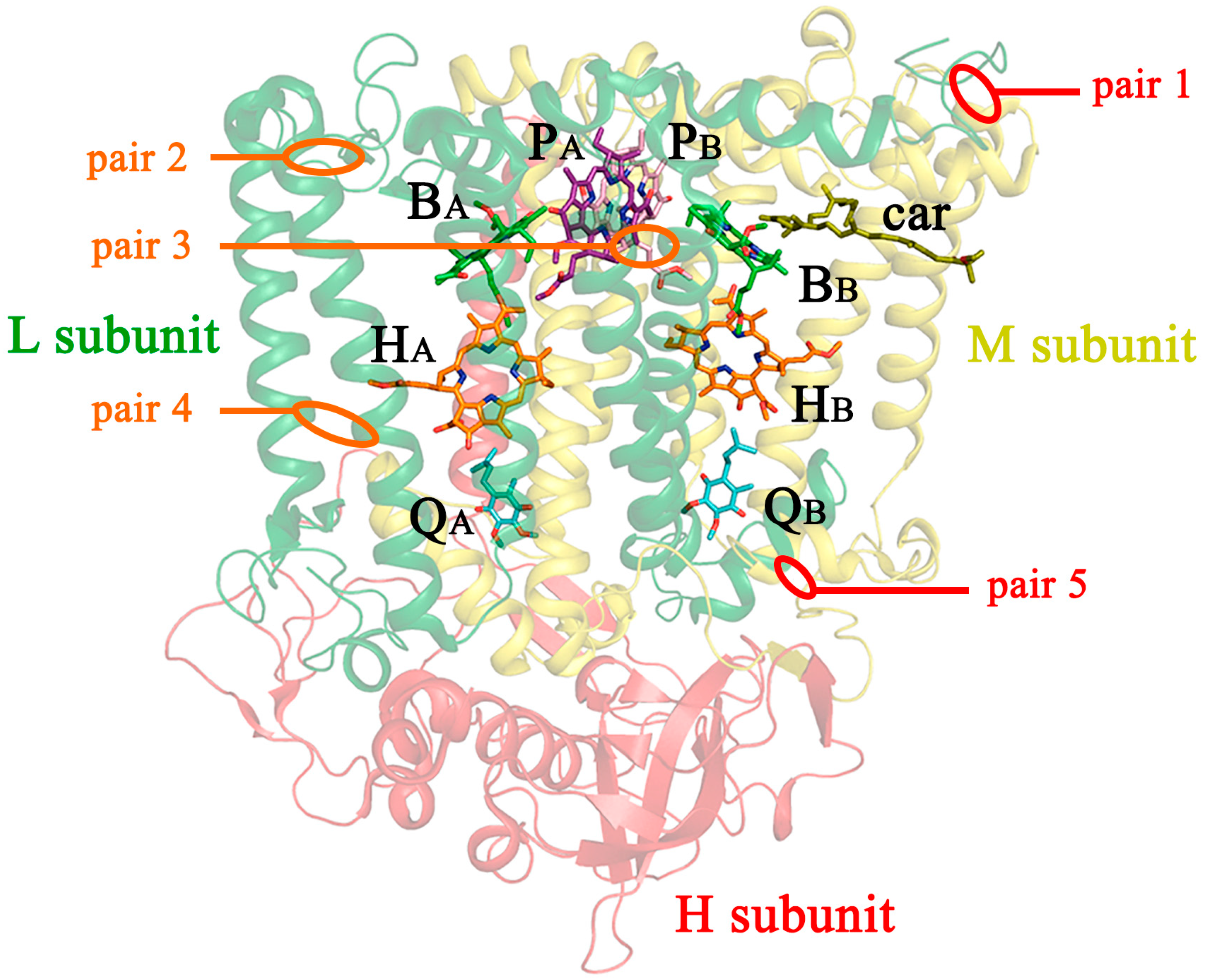
3.1. The Design and Introduction of Disulfide Bonds
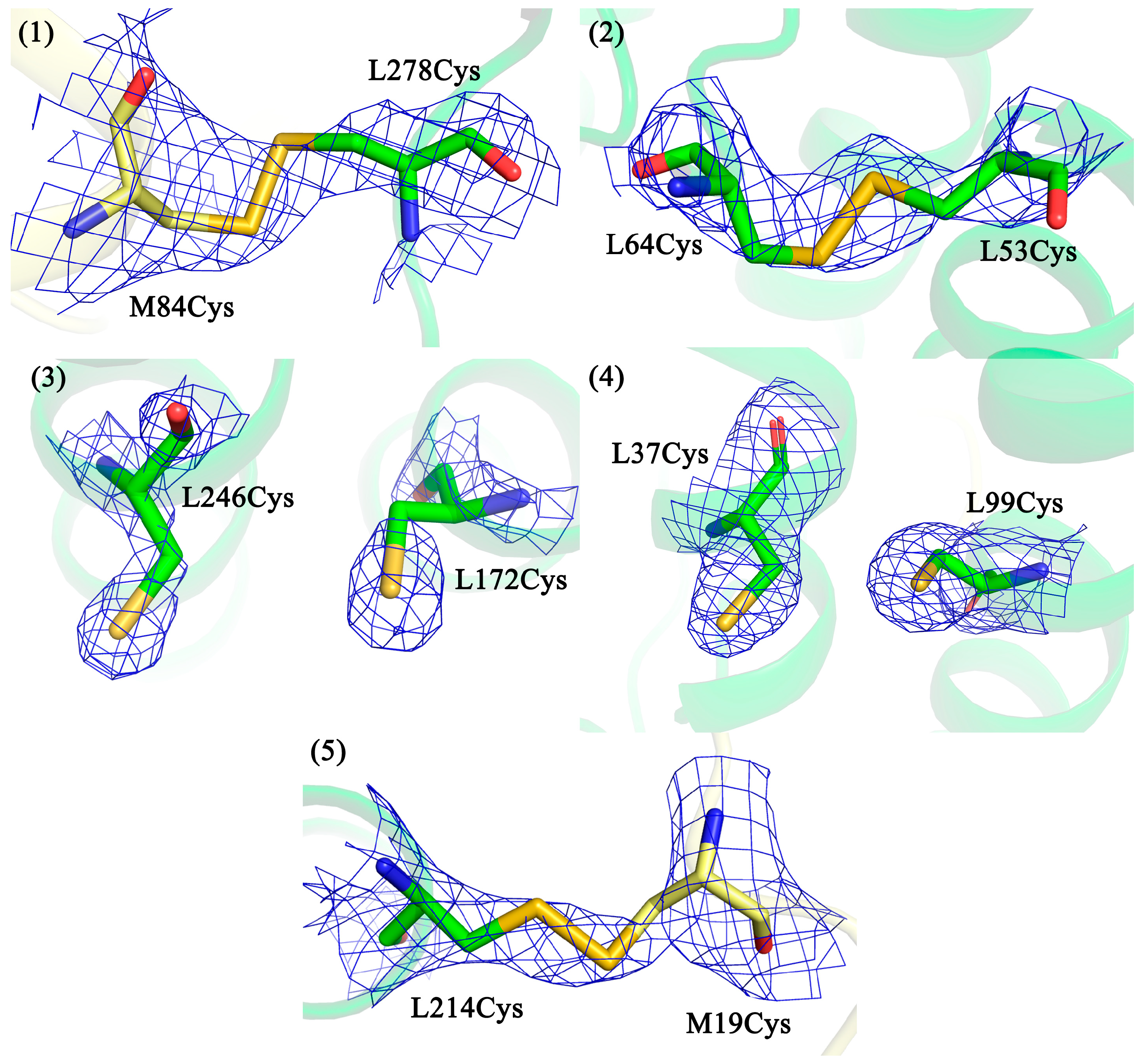
- (1)
- V(M84)C+G(L278)C, periplasmic surface, intra-subunit S-S bond;
- (2)
- A(L53)C+I(L64)C, periplasmic surface, inter-subunit S-S bond;
- (3)
- A(L172)C+L(L246)C, membrane zone closer to periplasm, inter-subunit S-S bond;
- (4)
- A(L37)C+S(L99)C, hydrophobic zone near BPheo HA, inter-subunit S-S bond;
- (5)
- G(M19)C+T(L214)C, cytoplasmic surface, intra-subunit S-S bond.
3.2. Confirmation of Disulfide Bond Formation by X-ray Crystallography
3.3. Confirmation of the Formation of Disulfide Bonds by Polyacrylamide Gel Electrophoresis (PAGE)
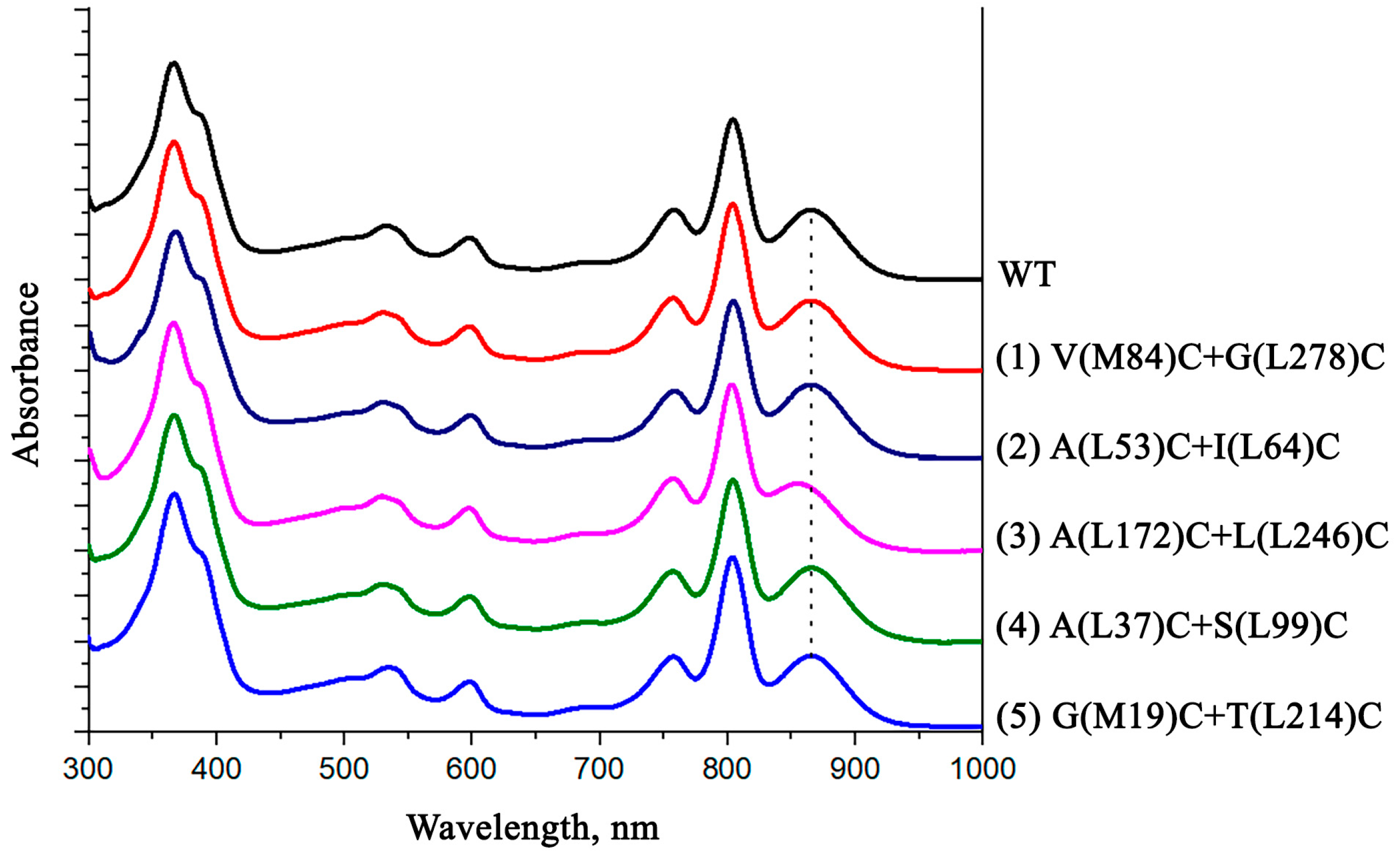
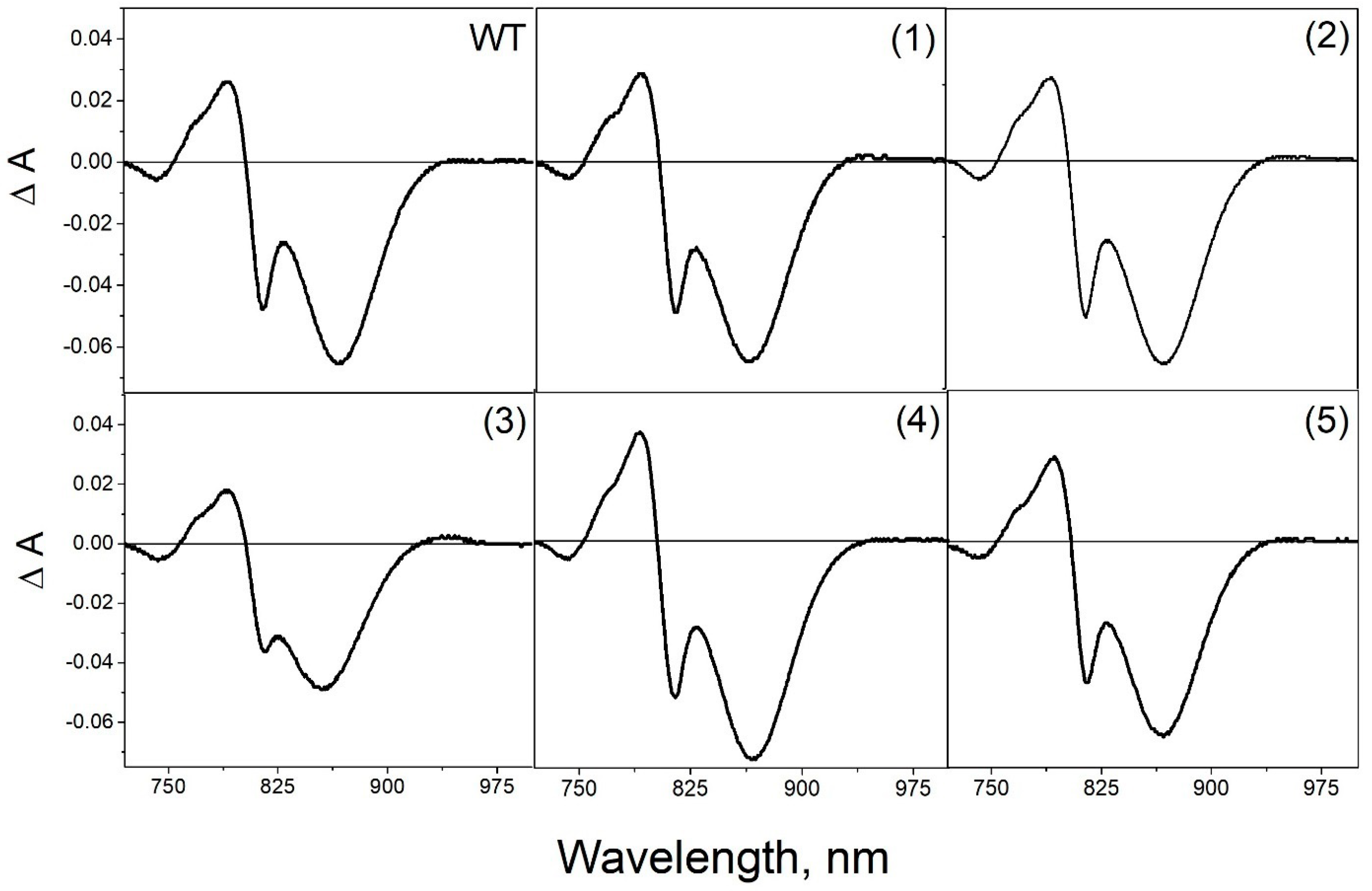
3.4. Pigment Content and Photochemical Properties of Mutant RCs
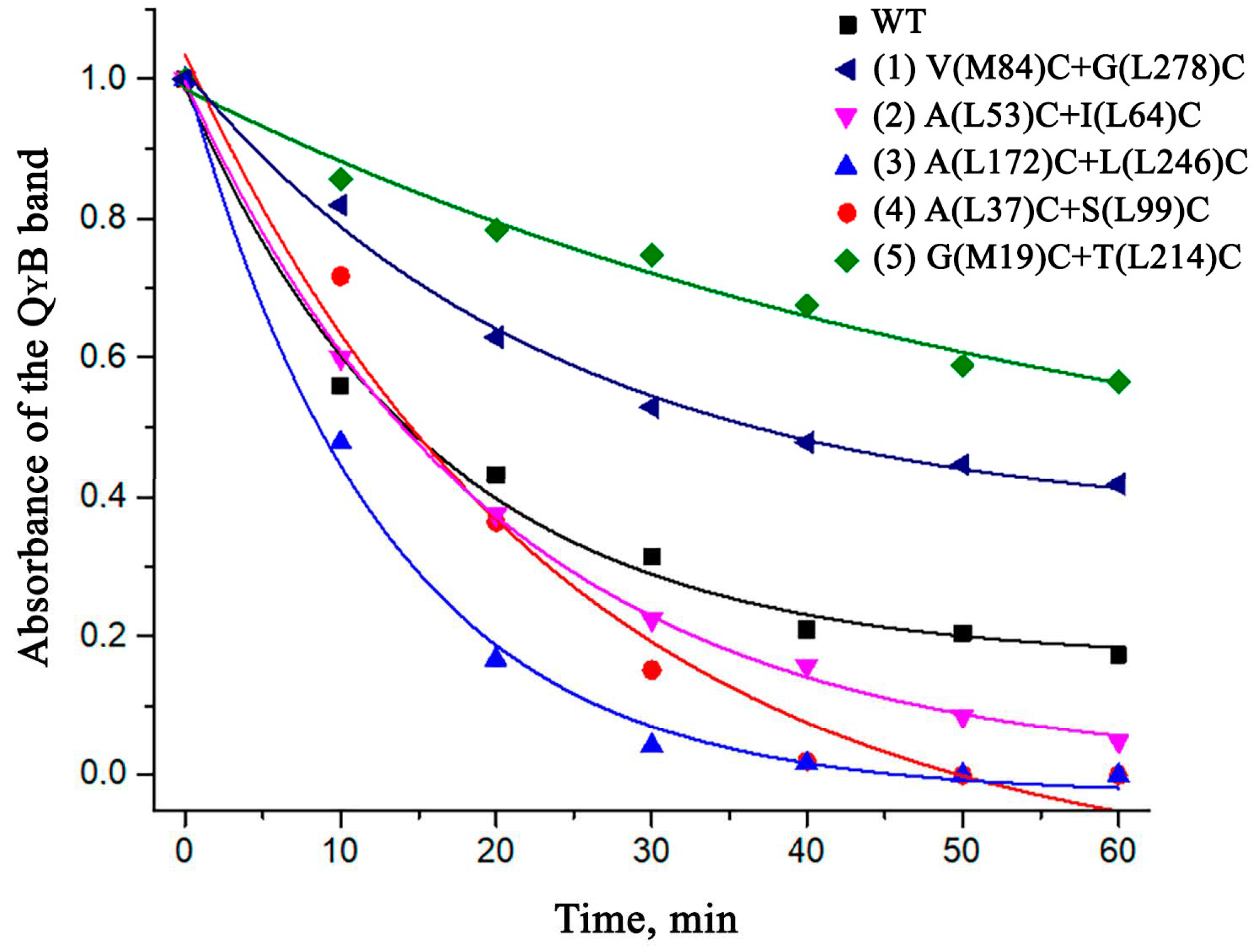
3.5. Thermostability of the Mutant RCs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, J.P.; Feher, G.; Yeates, T.O.; Komiya, H.; Rees, D.C. Structure of the Reaction Center from Rhodobacter Sphaeroides R-26: The Cofactors. Proc. Natl. Acad. Sci. USA 1987, 84, 5730–5734. [Google Scholar] [CrossRef] [Green Version]
- Kálmán, L.; Williams, J.C.; Allen, J.P. Comparison of Bacterial Reaction Centers and Photosystem II. Photosynth. Res. 2008, 98, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Pakula, A.A.; Sauer, R.T. Genetic analysis of protein stability and function. Annu. Rev. Genet. 1989, 23, 289–310. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A.; Gavira, J.A. Introduction to Protein Crystallization. Acta Cryst. F 2014, 70, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Wraight, C.A.; Clayton, R.K. The Absolute Quantum Efficiency of Bacteriochlorophyll Photooxidation in Reaction Centres of Rhodopseudomonas Spheroides. Biochim. Biophys. Acta BBA–Bioenerg. 1974, 333, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Grattieri, M.; Beaver, K.; Gaffney, E.M.; Dong, F.; Minteer, S.D. Advancing the Fundamental Understanding and Practical Applications of Photo-Bioelectrocatalysis. Chem. Commun. 2020, 56, 8553–8568. [Google Scholar] [CrossRef] [PubMed]
- Fufina, T.Y.; Vasilieva, L.G. Effect of Detergents and Osmolytes on Thermal Stability of Native and Mutant Rhodobacter Sphaeroides Reaction Centers. Biochem. Mosc. 2021, 86, 517–524. [Google Scholar] [CrossRef]
- Selikhanov, G.; Fufina, T.; Guenther, S.; Meents, A.; Gabdulkhakov, A.; Vasilieva, L. X-ray Structure of the Rhodobacter Sphaeroides Reaction Center with an M197 Phe→His Substitution Clarifies the Properties of the Mutant Complex. IUCrJ 2022, 9, 261–271. [Google Scholar] [CrossRef]
- Holden-Dye, K.; Crouch, L.I.; Williams, C.M.; Bone, R.A.; Cheng, J.; Böhles, F.; Heathcote, P.; Jones, M.R. Opposing Structural Changes in Two Symmetrical Polypeptides Bring about Opposing Changes to the Thermal Stability of a Complex Integral Membrane Protein. Arch. Biochem. Biophys. 2011, 505, 160–170. [Google Scholar] [CrossRef]
- Kangur, L.; Jones, M.R.; Freiberg, A. Hydrogen Bonds in the Vicinity of the Special Pair of the Bacterial Reaction Center Probed by Hydrostatic High-Pressure Absorption Spectroscopy. Biophys. Chem. 2017, 231, 27–33. [Google Scholar] [CrossRef]
- Jones, M.R.; Visschers, R.W.; Van Grondelle, R.; Hunter, C.N. Construction and Characterization of a Mutant of Rhodobacter Sphaeroides with the Reaction Center as the Sole Pigment-Protein Complex. Biochemistry 1992, 31, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Vasilieva, L.G.; Bolgarina, T.I.; Khatynov, R.A.; Shkuropatov, A.Y.; Miyake, J.; Shuvalov, V.A. Substitution of Valine-157 Residue by Tyrosine in the L-Subunit of the Rhodobacter Sphaeroides Reaction Center. Dokl. Biochem. Biophys. 2001, 376, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Naismith, J.H. An Efficient One-Step Site-Directed Deletion, Insertion, Single and Multiple-Site Plasmid Mutagenesis Protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatypov, R.A.; Vasilieva, L.G.; Fufina, T.Y.; Bolgarina, T.I.; Shuvalov, V.A. Substitution of Isoleucine L177 by Histidine Affects the Pigment Composition and Properties of the Reaction Center of the Purple Bacterium Rhodobacter Sphaeroides. Biochemistry 2005, 70, 1256–1261. [Google Scholar] [CrossRef]
- Fufina, T.Y.; Vasilieva, L.G.; Khatypov, R.A.; Shkuropatov, A.Y.; Shuvalov, V.A. Substitution of Isoleucine L177 by Histidine in Rhodobacter Sphaeroides Reaction Center Results in the Covalent Binding of PA Bacteriochlorophyll to the L Subunit. FEBS Lett. 2007, 581, 5769–5773. [Google Scholar] [CrossRef] [Green Version]
- Goldsmith, J.O.; Boxer, S.G. Rapid Isolation of Bacterial Photosynthetic Reaction Centers with an Engineered Poly-Histidine Tag. Biochim. Biophys. Acta BBA–Bioenerg. 1996, 1276, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Okamura, M.Y.; Steiner, L.A.; Feher, G. Characterization of Reaction Centers from Photosynthetic Bacteria. I. Subunit Structure of the Protein Mediating the Primary Photochemistry in Rhodopseudomonas Spheroides R-26. Biochemistry 1974, 13, 1394–1403. [Google Scholar] [CrossRef]
- Vasilieva, L.G.; Fufina, T.Y.; Gabdulkhakov, A.G.; Leonova, M.M.; Khatypov, R.A.; Shuvalov, V.A. The Site-Directed Mutation I(L177)H in Rhodobacter Sphaeroides Reaction Center Affects Coordination of PA and BB Bacteriochlorophylls. Biochim. Biophys. Acta BBA–Bioenerg. 2012, 1817, 1407–1417. [Google Scholar] [CrossRef]
- Fufina, T.Y.; Vasilieva, L.G.; Shuvalov, V.A. Examination of Stability of Mutant Photosynthetic Reaction Center of Rhodobacter Sphaeroides I(L177)H and Determination of Location of Bacteriochlorophyll Covalently Bound to the Protein. Biochem. Mosc. 2010, 75, 208–213. [Google Scholar] [CrossRef]
- Kashino, Y.; Koike, H.; Satoh, K. An Improved Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis System for the Analysis of Membrane Protein Complexes. Electrophoresis 2001, 22, 1004–1007. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gabdulkhakov, A.G.; Fufina, T.Y.; Vasilieva, L.G.; Mueller, U.; Shuvalov, V.A. Expression, Purification, Crystallization and Preliminary X-Ray Structure Analysis of Wild-Type and L(M196)H-Mutant Rhodobacter Sphaeroides Reaction Centres. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 506–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selikhanov, G.; Fufina, T.; Vasilieva, L.; Betzel, C.; Gabdulkhakov, A. Novel Approaches for the Lipid Sponge Phase Crystallization of the Rhodobacter Sphaeroides Photosynthetic Reaction Center. IUCrJ 2020, 7, 1084–1091. [Google Scholar] [CrossRef]
- Nurizzo, D.; Mairs, T.; Guijarro, M.; Rey, V.; Meyer, J.; Fajardo, P.; Chavanne, J.; Biasci, J.-C.; McSweeney, S.; Mitchell, E. The ID23-1 Structural Biology Beamline at the ESRF. J. Synchrotron Radiat. 2006, 13, 227–238. [Google Scholar] [CrossRef]
- Gabadinho, J.; Beteva, A.; Guijarro, M.; Rey-Bakaikoa, V.; Spruce, D.; Bowler, M.W.; Brockhauser, S.; Flot, D.; Gordon, E.J.; Hall, D.R.; et al. MxCuBE: A Synchrotron Beamline Control Environment Customized for Macromolecular Crystallography Experiments. J. Synchrotron Radiat. 2010, 17, 700–707. [Google Scholar] [CrossRef]
- Bourenkov, G.P.; Popov, A.N. Optimization of Data Collection Taking Radiation Damage into Account. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. XDS . Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC 5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System. 2022. Available online: http://www.pymol.org (accessed on 8 November 2022).
- Netto, L.E.S.; de Oliveira, M.A.; Monteiro, G.; Demasi, A.P.D.; Cussiol, J.R.R.; Discola, K.F.; Demasi, M.; Silva, G.M.; Alves, S.V.; Faria, V.G.; et al. Reactive Cysteine in Proteins: Protein Folding, Antioxidant Defense, Redox Signaling and More. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 146, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Reiss, B.D.; Hanson, D.K.; Firestone, M.A. Evaluation of the Photosynthetic Reaction Center Protein for Potential Use as a Bioelectronic Circuit Element. Biotechnol. Prog. 2007, 23, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent Binding Explains Anomalous SDS-PAGE Migration of Membrane Proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [Green Version]
- Clayton, R.K.; Haselkorn, R. Protein Components of Bacterial Photosynthetic Membranes. J. Mol. Biol. 1972, 68, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, W.D.; Kaplan, S. Effect of Heat and 2-Mercaptoethanol on Intracytoplasmic Membrane Polypeptides of Rhodopseudomonas Sphaeroides. J. Bacteriol. 1978, 135, 656–667. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-X.; Li, L.; Yang, X.-Z.; Wei, C.-W.; Cheng, H.-M.; Gao, S.-Q.; Wen, G.-B.; Lin, Y.-W. Enhancement of Protein Stability by an Additional Disulfide Bond Designed in Human Neuroglobin. RSC Adv. 2019, 9, 4172–4179. [Google Scholar] [CrossRef] [Green Version]
- van Beek, H.L.; Wijma, H.J.; Fromont, L.; Janssen, D.B.; Fraaije, M.W. Stabilization of Cyclohexanone Monooxygenase by a Computationally Designed Disulfide Bond Spanning Only One Residue. FEBS Open Biol. 2014, 4, 168–174. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Romero, I.; Ariza, A.; Wilson, K.S.; Skjøt, M.; Vind, J.; De Maria, L.; Skov, L.K.; Sanchez-Ruiz, J.M. Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks. PLoS ONE 2013, 8, e70013. [Google Scholar] [CrossRef] [Green Version]
- Creighton, T.E.; Zapun, A.; Darby, N.J. Mechanisms and Catalysts of Disulphide Bond Formation in Proteins. Trends Biotechnol. 1995, 13, 18–23. [Google Scholar] [CrossRef]
- Fass, D. Disulfide Bonding in Protein Biophysics. Annu. Rev. Biophys. 2012, 41, 63–79. [Google Scholar] [CrossRef]
- Dill, K.A. Dominant Forces in Protein Folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Cowburn, D. Combining Biophysical Methods to Analyze the Disulfide Bond in SH2 Domain of C-Terminal Src Kinase. Biophys. Rep. 2016, 2, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, M.; Signor, G.; Matthews, B.W. Substantial Increase of Protein Stability by Multiple Disulphide Bonds. Nature 1989, 342, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Gross, A.K.; Oprian, D.D. An Opsin Mutant with Increased Thermal Stability. Biochemistry 2003, 42, 1995–2001. [Google Scholar] [CrossRef]
- Standfuss, J.; Xie, G.; Edwards, P.C.; Burghammer, M.; Oprian, D.D.; Schertler, G.F.X. Crystal Structure of a Thermally Stable Rhodopsin Mutant. J. Mol. Biol. 2007, 372, 1179–1188. [Google Scholar] [CrossRef] [Green Version]
- Popov, P.; Peng, Y.; Shen, L.; Stevens, R.C.; Cherezov, V.; Liu, Z.-J.; Katritch, V. Computational Design of Thermostabilizing Point Mutations for G Protein-Coupled Receptors. eLife 2018, 7, e34729. [Google Scholar] [CrossRef]
- Landeta, C.; Boyd, D.; Beckwith, J. Disulfide Bond Formation in Prokaryotes. Nat. Microbiol. 2018, 3, 270–280. [Google Scholar] [CrossRef]
- Grattieri, M.; Minteer, S.D. Self-Powered Biosensors. ACS Sens. 2018, 3, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Kornienko, N.; Zhang, J.Z.; Sakimoto, K.K.; Yang, P.; Reisner, E. Interfacing Nature’s Catalytic Machinery with Synthetic Materials for Semi-Artificial Photosynthesis. Nat. Nanotech. 2018, 13, 890–899. [Google Scholar] [CrossRef]
- Grattieri, M. Purple Bacteria Photo-Bioelectrochemistry: Enthralling Challenges and Opportunities. Photochem. Photobiol. Sci. 2020, 19, 424–435. [Google Scholar] [CrossRef]
- Hasan, K.; Grippo, V.; Sperling, E.; Packer, M.A.; Leech, D.; Gorton, L. Evaluation of Photocurrent Generation from Different Photosynthetic Organisms. ChemElectroChem 2017, 4, 412–417. [Google Scholar] [CrossRef]
- Milano, F.; Punzi, A.; Ragni, R.; Trotta, M.; Farinola, G.M. Photonics and Optoelectronics with Bacteria: Making Materials from Photosynthetic Microorganisms. Adv. Funct. Mater. 2019, 29, 1805521. [Google Scholar] [CrossRef]
- Kruse, O.; Rupprecht, J.; Mussgnug, J.H.; Dismukes, G.C.; Hankamer, B. Photosynthesis: A Blueprint for Solar Energy Capture and Biohydrogen Production Technologies. Photochem. Photobiol. Sci. 2005, 4, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Almén, M.S.; Nordström, K.J.; Fredriksson, R.; Schiöth, H.B. Mapping the Human Membrane Proteome: A Majority of the Human Membrane Proteins Can Be Classified According to Function and Evolutionary Origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef] [PubMed]

| L37Cys+L99Cys | L53Cys+L64Cys | L172Cys+L246Cys | M19Cys+L214Cys | M84Cys+L278Cys | |
|---|---|---|---|---|---|
| Diffraction source | ESRF, beamline ID30A-3 | Proteum X8 (Bruker) | XtaLAB Synergy-S (Rigaku) | ESRF, beamline ID30A-3 | ESRF, beamline ID30A-3 |
| Wavelength (Å) | 0.9677 | 1.54178 | 1.54178 | 0.9677 | 0.9677 |
| Temperature (K) | 100 | 110 | 120 | 100 | 100 |
| Detector | DECTRIS Eiger X 4M | PLATINUM135 CCD | HyPix-6000C | DECTRIS Eiger X 4M | DECTRIS Eiger X 4M |
| Rotation range per image (°) | 0.15 | 0.5 | 0.5 | 0.05 | 0.1 |
| Total rotation range (°) | 150 | 180 | 120 | 50 | 200 |
| Space group | P3121 | P3121 | P41212 | P41212 | C2 |
| a, b, c (Å), α, β, γ, ◦ | 139.8 139.8 186.5 90 90 120 | 139.6 139.6 185.0 90 90 120 | 99.7 99.7 239.1 90 90 90 | 100.91 100.91 237.0 90 90 90 | 253.1 75.9 65.8 90.0 95.5 90.0 |
| Resolution range (Å) | 30.00–2.60 (2.67–2.60) | 30.00–2.85 (2.95–2.85) | 30.00–2.30 (2.40–2.30) | 30.00–2.75 (2.82–2.75) | 30.00–2.60 (2.67–2.60) |
| Total No. of reflections | 564 510 (43 474) | 283 148 (16 729) | 366 770 (30 085) | 115 943 (8 973) | 150 052 (11 638) |
| No. of unique reflections | 65 303 (4 784) | 49 267 (4 765) | 53 136 (5 315) | 32 119 (2 369) | 38 037 (2 832) |
| Completeness (%) | 99.9 (100.0) | 99.7 (97.6) | 97.3 (82.7) | 98.1 (99.9) | 98.7 (99.3) |
| Redundancy | 8.6 (9.1) | 5.7 (3.5) | 6.9 (5.7) | 3.6 (3.8) | 3.9 (4.1) |
| 〈I/σ(I)〉 | 10.13 (1.42) | 5.42 (1.79) | 8.26 (1.10) | 7.92 (1.06) | 8.15 (1.02) |
| Rr.i.m.‡ | 17.3 (161.4) | 27.1 (58.2) | 11.7 (65.8) | 11.9 (124.4) | 9.8 (134.9) |
| CC1/2 | 99.7 (57.9) | 99.6 (59.4) | 96.6 (60.2) | 99.6 (40.1) | 99.7 (58.6) |
| L37Cys+L99Cys | L53Cys+L64Cys | L172Cys+L246Cys | M19Cys+L214Cys | M84Cys+L278Cys | |
|---|---|---|---|---|---|
| Resolution range (Å) | 30.00–2.60 (2.64–2.60) | 30.00–2.85 (2.95–2.85) | 30.00–2.30 (2.40–2.30) | 46.0–2.75 (2.82–2.75) | 41.00–2.60 (2.67–2.60) |
| Completeness (%) | 99.9 (100.0) | 99.5 (96.1) | 99.3 (99.0) | 98.0 (96.0) | 98.6 (99.0) |
| No. of reflections, working set | 65 301 (2 567) | 48 500 (2 565) | 44 919 (2 614) | 32 091 (3 403) | 38 001 (3 296) |
| No. of reflections, test set | 3 351 (153) | 2 435 (131) | 2 273 (140) | 1 282 (141) | 1 519 (138) |
| Rcryst | 18.56 (25.95) | 22.85 (33.77) | 25.86 (26.08) | 19.91 (34.63) | 19.27 (40.40) |
| Rfree | 20.74 (30.34) | 27.90 (38.49) | 30.82 (32.56) | 27.06 (42.24) | 24.93 (40.01) |
| R.m.s. deviations | |||||
| Bonds (Å) | 0.009 | 0.010 | 0.010 | 0.010 | 0.009 |
| Angles (°) | 1.122 | 1.314 | 1.354 | 1.234 | 1.159 |
| PDB ID | 8C5X | 8C6K | 8C87 | 8C88 | 8C7C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selikhanov, G.; Atamas, A.; Yukhimchuk, D.; Fufina, T.; Vasilieva, L.; Gabdulkhakov, A. Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds. Membranes 2023, 13, 154. https://doi.org/10.3390/membranes13020154
Selikhanov G, Atamas A, Yukhimchuk D, Fufina T, Vasilieva L, Gabdulkhakov A. Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds. Membranes. 2023; 13(2):154. https://doi.org/10.3390/membranes13020154
Chicago/Turabian StyleSelikhanov, Georgii, Anastasia Atamas, Diana Yukhimchuk, Tatiana Fufina, Lyudmila Vasilieva, and Azat Gabdulkhakov. 2023. "Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds" Membranes 13, no. 2: 154. https://doi.org/10.3390/membranes13020154
APA StyleSelikhanov, G., Atamas, A., Yukhimchuk, D., Fufina, T., Vasilieva, L., & Gabdulkhakov, A. (2023). Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds. Membranes, 13(2), 154. https://doi.org/10.3390/membranes13020154







