DNA Transactions in Bacteria and Membranes: A Place for the Hfq Protein?
Abstract
1. Introduction
2. Materials and Methods
3. Mechanisms of DNA-Mediated Membrane Interaction
3.1. Direct Versus Indirect Membrane–DNA Interaction
3.2. Methods to Analyze Membrane–DNA Interaction
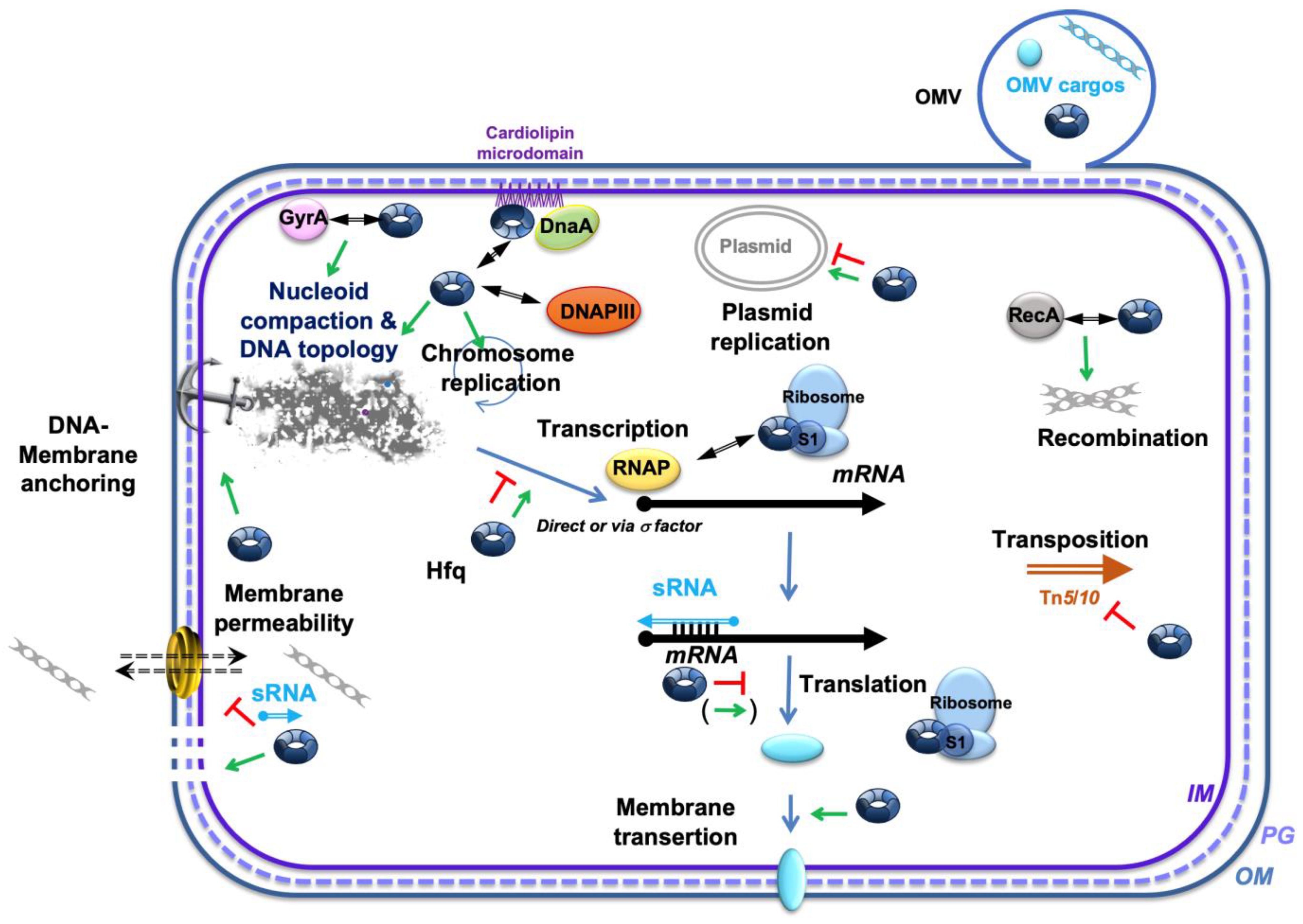
4. Regulatory Processes Related to the Importance of Membranes in Different DNA Transactions and the Potential Role of Hfq
4.1. DNA Replication
4.2. Genetic Recombination, DNA Damage/Mutagenesis and Repair
4.3. Other DNA Transactions: Transposition, OMV DNA Cargo-Loading, Nucleoid Organization
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| sRNA | Small noncoding RNA |
| IM/OM | Inner/outer membrane |
| NAP | Nucleoid-Associated Protein |
| RNAP/DNAP | RNA or DNA polymerase |
| Tn | Transposon |
| OMV | Outer membrane vesicles |
| LLPS | Liquid–Liquid Phase Separation |
| PTM | Post-translational modification |
| SRP | Signal Recognition Particle |
References
- Joyeux, M. A segregative phase separation scenario of the formation of the bacterial nucleoid. Soft Matter 2018, 14, 7368–7381. [Google Scholar] [PubMed]
- Stonington, O.G.; Pettijohn, D.E. The folded genome of Escherichia coli isolated in a protein-DNA-RNA complex. Proc. Natl. Acad. Sci. USA 1971, 68, 6–9. [Google Scholar] [CrossRef]
- Worcel, A.; Burgi, E. Properties of a membrane-attached form of the folded chromosome of Escherichia coli. J. Mol. Biol. 1974, 82, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Sinden, R.R. DNA Structure and Function; Academic Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Kuzminov, A. Bacterial nucleoid is a riddle wrapped in a mystery inside an enigma. J. Bacteriol. 2024, 206, e00211-23. [Google Scholar] [CrossRef]
- Verma, S.C.; Qian, Z.; Adhya, S.L. Architecture of the Escherichia coli nucleoid. PLoS Genet. 2019, 15, e1008456. [Google Scholar] [CrossRef]
- Dworsky, P. Membrane attachment of folded chromosome of Escherichia coli. Biochem. J. 1976, 154, 239–241. [Google Scholar] [CrossRef]
- Roggiani, M.; Goulian, M. Chromosome-Membrane Interactions in Bacteria. Annu. Rev. Genet. 2015, 49, 115–129. [Google Scholar] [CrossRef]
- Cossa, A.; Trepout, S.; Wien, F.; Groen, J.; Le Brun, E.; Turbant, F.; Besse, L.; Pereiro, E.; Arluison, V. Cryo soft X-ray tomography to explore Escherichia coli nucleoid remodeling by Hfq master regulator. J. Struct. Biol. 2022, 214, 107912. [Google Scholar] [CrossRef]
- Morgan, C.; Rosenkranz, H.S.; Carr, H.S.; Rose, H.M. Electron microscopy of chloramphenicol-treated Escherichia coli. J. Bacteriol. 1967, 93, 1987–2002. [Google Scholar] [CrossRef]
- Dubochet, J.; McDowall, A.W.; Menge, B.; Schmid, E.N.; Lickfeld, K.G. Electron microscopy of frozen-hydrated bacteria. J. Bacteriol. 1983, 155, 381–390. [Google Scholar] [CrossRef]
- Pilhofer, M.; Ladinsky, M.S.; McDowall, A.W.; Jensen, G.J. Bacterial TEM: New insights from cryo-microscopy. Methods Cell Biol. 2010, 96, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Cossa, A.; Wien, F.; Turbant, F.; Kaczorowski, T.; Wegrzyn, G.; Arluison, V.; Perez-Berna, A.J.; Trepout, S.; Pereiro, E. Evaluation of the Role of Bacterial Amyloid on Nucleoid Structure Using Cryo-Soft X-Ray Tomography. Methods Mol. Biol. 2022, 2538, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Sekimizu, K.; Kornberg, A. Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 1988, 263, 7131–7135. [Google Scholar] [PubMed]
- Bisicchia, P.; Steel, B.; Mariam Debela, M.H.; Lowe, J.; Sherratt, D. The N-terminal membrane-spanning domain of the Escherichia coli DNA translocase FtsK hexamerizes at midcell. mBio 2013, 4, e00800-13. [Google Scholar] [CrossRef]
- Cech, G.M.; Szalewska-Palasz, A.; Kubiak, K.; Malabirade, A.; Grange, W.; Arluison, V.; Wegrzyn, G. The Escherichia Coli Hfq Protein: An Unattended DNA-Transactions Regulator. Front. Mol. Biosci. 2016, 3, 36. [Google Scholar] [CrossRef]
- Franze de Fernandez, M.T.; Hayward, W.S.; August, J.T. Bacterial proteins required for replication of phage Qb ribonucleic acid. J. Biol. Chem. 1972, 247, 824–831. [Google Scholar] [CrossRef]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Sun, X.; Zhulin, I.; Wartell, R.M. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002, 30, 3662–3671. [Google Scholar]
- Feliciano, J.R.; Seixas, A.M.M.; Pita, T.; Leitao, J.H. Comparative Genomics and Evolutionary Analysis of RNA-Binding Proteins of Burkholderia cenocepacia J2315 and Other Members of the B. cepacia Complex. Genes 2020, 11, 231. [Google Scholar] [CrossRef]
- Bouloc, P.; Repoila, F. Fresh layers of RNA-mediated regulation in Gram-positive bacteria. Curr. Opin. Microbiol. 2016, 30, 30–35. [Google Scholar] [CrossRef]
- Gottesman, S.; McCullen, C.A.; Guillier, M.; Vanderpool, C.K.; Majdalani, N.; Benhammou, J.; Thompson, K.M.; FitzGerald, P.C.; Sowa, N.A.; FitzGerald, D.J. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 1–11. [Google Scholar] [PubMed]
- Yao, H.; Kang, M.; Wang, Y.; Feng, Y.; Kong, S.; Cai, X.; Ling, Z.; Chen, S.; Jiao, X.; Yin, Y. An essential role for hfq involved in biofilm formation and virulence in serotype 4b Listeria monocytogenes. Microbiol. Res. 2018, 215, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar]
- Geinguenaud, F.; Calandrini, V.; Teixeira, J.; Mayer, C.; Liquier, J.; Lavelle, C.; Arluison, V. Conformational transition of DNA bound to Hfq probed by infrared spectroscopy. Phys. Chem. Chem. Phys. 2011, 13, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, K.; Wien, F.; Yadav, I.; Jones, N.C.; Vrønning Hoffmann, S.; Le Cam, E.; Cossa, A.; Geinguenaud, F.; van der Maarel, J.R.C.; Węgrzyn, G.; et al. Amyloid-like Hfq interaction with single-stranded DNA: Involvement in recombination and replication in Escherichia coli. QRB Discov. 2022, 3, e15. [Google Scholar] [CrossRef]
- Wien, F.; Gragera, M.; Matsuo, T.; Moroy, G.; Bueno-Carrasco, M.T.; Arranz, R.; Cossa, A.; Martel, A.; Bordallo, H.N.; Rudic, S.; et al. Amyloid-like DNA bridging: A new mode of DNA shaping. Nucleic Acids Res. 2025, 53, gkaf169. [Google Scholar] [CrossRef]
- Diestra, E.; Cayrol, B.; Arluison, V.; Risco, C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS ONE 2009, 4, e8301. [Google Scholar] [CrossRef]
- Caillet, J.; Baron, B.; Boni, I.V.; Caillet-Saguy, C.; Hajnsdorf, E. Identification of protein-protein and ribonucleoprotein complexes containing Hfq. Sci. Rep. 2019, 9, 14054. [Google Scholar] [CrossRef]
- Brennan, R.G.; Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007, 10, 125–133. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Pearson, R.F.; Moller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: A bacterial Sm-like protein. EMBO J. 2002, 21, 3546–3556. [Google Scholar]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef] [PubMed]
- Updegrove, T.B.; Correia, J.J.; Galletto, R.; Bujalowski, W.; Wartell, R.M. E. coli DNA associated with isolated Hfq interacts with Hfq’s distal surface and C-terminal domain. Biochim. Biophys. Acta 2010, 1799, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Orans, J.; Kovach, A.R.; Hoff, K.E.; Horstmann, N.M.; Brennan, R.G. Crystal structure of an Escherichia coli Hfq Core (residues 2-69)-DNA complex reveals multifunctional nucleic acid binding sites. Nucleic Acids Res. 2020, 48, 3987–3997. [Google Scholar] [PubMed]
- Dimastrogiovanni, D.; Frohlich, K.S.; Bandyra, K.J.; Bruce, H.A.; Hohensee, S.; Vogel, J.; Luisi, B.F. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife 2014, 3, e05375. [Google Scholar] [CrossRef]
- Olsen, A.S.; Moller-Jensen, J.; Brennan, R.G.; Valentin-Hansen, P. C-Terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010, 404, 173–182. [Google Scholar] [CrossRef]
- Arluison, V.; Folichon, M.; Marco, S.; Derreumaux, P.; Pellegrini, O.; Seguin, J.; Hajnsdorf, E.; Regnier, P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004, 271, 1258–1265. [Google Scholar]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trepout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35, e00190. [Google Scholar] [CrossRef]
- Berbon, M.; Martinez, D.; Morvan, E.; Grelard, A.; Kauffmann, B.; Waeytens, J.; Wien, F.; Arluison, V.; Habenstein, B. Hfq C-terminal region forms a beta-rich amyloid-like motif without perturbing the N-terminal Sm-like structure. Commun. Biol. 2023, 6, 1075. [Google Scholar] [CrossRef]
- Malabirade, A.; Morgado-Brajones, J.; Trepout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 10724. [Google Scholar] [CrossRef]
- Turbant, F.; Waeytens, J.; Campidelli, C.; Bombled, M.; Martinez, D.; Grelard, A.; Habenstein, B.; Raussens, V.; Velez, M.; Wien, F.; et al. Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq. Int. J. Mol. Sci. 2022, 23, 8739. [Google Scholar] [CrossRef]
- Turbant, F.; Blache, A.; Wegrzyn, G.; Achouak, W.; Wien, F.; Arluison, V. Use of Synchrotron Radiation Circular Dichroism to Analyze the Interaction and Insertion of Proteins into Bacterial Outer Membrane Vesicles. Methods Mol. Biol. 2024, 2843, 73–94. [Google Scholar] [CrossRef] [PubMed]
- Turbant, F.; Wu, P.; Wien, F.; Arluison, V. The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology 2021, 10, 900. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Hara, H.; Fishov, I.; Mileykovskaya, E.; Norris, V. The membrane: Transertion as an organizing principle in membrane heterogeneity. Front. Microbiol. 2015, 6, 572. [Google Scholar] [CrossRef]
- Pramanik, J.; Keasling, J.D. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 1997, 56, 398–421. [Google Scholar] [CrossRef]
- Card, G.L.; Trautman, J.K. Role of anionic lipid in bacterial membranes. Biochim. Biophys. Acta 1990, 1047, 77–82. [Google Scholar] [CrossRef]
- Asif, A.; Mohsin, H.; Tanvir, R.; Rehman, Y. Revisiting the Mechanisms Involved in Calcium Chloride Induced Bacterial Transformation. Front. Microbiol. 2017, 8, 2169. [Google Scholar] [CrossRef]
- Lloubes, R.; Cascales, E.; Walburger, A.; Bouveret, E.; Lazdunski, C.; Bernadac, A.; Journet, L. The Tol-Pal proteins of the Escherichia coli cell envelope: An energized system required for outer membrane integrity? Res. Microbiol. 2001, 152, 523–529. [Google Scholar] [CrossRef]
- Cohen-Khait, R.; Harmalkar, A.; Pham, P.; Webby, M.N.; Housden, N.G.; Elliston, E.; Hopper, J.T.S.; Mohammed, S.; Robinson, C.V.; Gray, J.J.; et al. Colicin-Mediated Transport of DNA through the Iron Transporter FepA. mBio 2021, 12, e0178721. [Google Scholar] [CrossRef]
- Adams, D.W.; Wu, L.J.; Errington, J. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 2015, 34, 491–501. [Google Scholar] [CrossRef]
- Sekimizu, K.; Yung, B.Y.; Kornberg, A. The dnaA protein of Escherichia coli. Abundance, improved purification, and membrane binding. J. Biol. Chem. 1988, 263, 7136–7140. [Google Scholar]
- Snider, J.; Kittanakom, S.; Damjanovic, D.; Curak, J.; Wong, V.; Stagljar, I. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat. Protoc. 2010, 5, 1281–1293. [Google Scholar] [CrossRef]
- Brenowitz, M.; Senear, D.F.; Shea, M.A.; Ackers, G.K. Quantitative DNase footprint titration: A method for studying protein-DNA interactions. Methods Enzymol. 1986, 130, 132–181. [Google Scholar] [CrossRef] [PubMed]
- Collas, P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010, 45, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Cava, D.G.; Velez, M. Supported Lipid Bilayers (SLBs) to Study Amyloid-Lipid Membrane Interactions with Atomic Force Microscopy. Methods Mol. Biol. 2022, 2538, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Cava, D.G.; Velez, M. Study of Amyloid Fibers Using Atomic Force Microscopy. Methods Mol. Biol. 2022, 2538, 1–11. [Google Scholar] [CrossRef]
- Waeytens, J.; Turbant, F.; Arluison, V.; Raussens, V.; Wien, F. Analysis of Bacterial Amyloid Interaction with Lipidic Membrane by Orientated Circular Dichroism and Infrared Spectroscopies. Methods Mol. Biol. 2022, 2538, 217–234. [Google Scholar] [CrossRef]
- Jacob, F.; Brenner, S. On the regulation of DNA synthesis in bacteria: The hypothesis of the replicon. C. R. Hebd. Seances Acad. Sci. 1963, 256, 298–300. [Google Scholar]
- Korn, D.; Thomas, M. Control of plasmid replication in Escherichia coli: Correlation of the membrane site of DNA replication with the bacterial segregation unit. Proc. Natl. Acad. Sci. USA 1971, 68, 2047–2051. [Google Scholar] [CrossRef]
- Winston, S.; Sueoka, N. DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1980, 77, 2834–2838. [Google Scholar] [CrossRef]
- Funnell, B.E. Participation of the bacterial membrane in DNA replication and chromosome partition. Trends Cell Biol. 1993, 3, 20–25. [Google Scholar] [CrossRef]
- Firshein, W.; Kim, P. Plasmid replication and partition in Escherichia coli: Is the cell membrane the key? Mol. Microbiol. 1997, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Arjes, H.A.; Kriel, A.; Sorto, N.A.; Shaw, J.T.; Wang, J.D.; Levin, P.A. Failsafe mechanisms couple division and DNA replication in bacteria. Curr. Biol. 2014, 24, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Rokop, M.E.; Auchtung, J.M.; Grossman, A.D. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 2004, 52, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.S.B.; Tran, N.T.; Wu, L.J.; Ramakrishnan, K.; Rejzek, M.; Gobbato, G.; Stevenson, C.E.M.; Lawson, D.M.; Errington, J.; Le, T.B.K. CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc. Mol. Cell 2021, 81, 3623–3636.e6. [Google Scholar] [CrossRef]
- Bryant, J.A.; Staunton, K.A.; Doherty, H.M.; Alao, M.B.; Ma, V.; Morcinek-Orłowska, J.; Goodall, E.C.; Gray, J.; Milner, M.; Cole, J.A.; et al. Bam complex associated proteins in Escherichia coli are functionally linked to peptidoglycan biosynthesis, membrane fluidity and DNA replication. eLife 2024, 13, RP99955. [Google Scholar] [CrossRef]
- Newman, G.; Crooke, E. DnaA, the initiator of Escherichia coli chromosomal replication, is located at the cell membrane. J. Bacteriol. 2000, 182, 2604–2610. [Google Scholar] [CrossRef]
- Regev, T.; Myers, N.; Zarivach, R.; Fishov, I. Association of the chromosome replication initiator DnaA with the Escherichia coli inner membrane in vivo: Quantity and mode of binding. PLoS ONE 2012, 7, e36441. [Google Scholar] [CrossRef]
- Crooke, E.; Hwang, D.S.; Skarstad, K.; Thony, B.; Kornberg, A.E. coli minichromosome replication: Regulation of initiation at oriC. Res. Microbiol. 1991, 142, 127–130. [Google Scholar] [CrossRef]
- Castuma, C.E.; Crooke, E.; Kornberg, A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J. Biol. Chem. 1993, 268, 24665–24668. [Google Scholar]
- Bramhill, D.; Kornberg, A. A model for initiation at origins of DNA replication. Cell 1988, 54, 915–918. [Google Scholar] [CrossRef]
- Yung, B.Y.; Kornberg, A. Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 7202–7205. [Google Scholar] [CrossRef] [PubMed]
- Boeneman, K.; Crooke, E. Chromosomal replication and the cell membrane. Curr. Opin. Microbiol. 2005, 8, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Fingland, N.; Patil, D.; Sharma, A.K.; Crooke, E. Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 2013, 14, 8517–8537. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.; Crooke, E. Membrane regulation of the chromosomal replication activity of E.coli DnaA requires a discrete site on the protein. EMBO J. 1996, 15, 2313–2321. [Google Scholar] [CrossRef]
- Aranovich, A.; Gdalevsky, G.Y.; Cohen-Luria, R.; Fishov, I.; Parola, A.H. Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J. Biol. Chem. 2006, 281, 12526–12534. [Google Scholar] [CrossRef]
- Garner, J.; Durrer, P.; Kitchen, J.; Brunner, J.; Crooke, E. Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J. Biol. Chem. 1998, 273, 5167–5173. [Google Scholar] [CrossRef]
- Patil, D.; Xun, D.; Schueritz, M.; Bansal, S.; Cheema, A.; Crooke, E.; Saxena, R. Membrane Stress Caused by Unprocessed Outer Membrane Lipoprotein Intermediate Pro-Lpp Affects DnaA and Fis-Dependent Growth. Front. Microbiol. 2021, 12, 677812. [Google Scholar] [CrossRef]
- Cech, G.M.; Pakula, B.; Kamrowska, D.; Wegrzyn, G.; Arluison, V.; Szalewska-Palasz, A. Hfq protein deficiency in Escherichia coli affects ColE1-like but not lambda plasmid DNA replication. Plasmid 2014, 73, 10–15. [Google Scholar] [CrossRef]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef]
- Gaffke, L.; Kubiak, K.; Cyske, Z.; Wegrzyn, G. Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics. Int. J. Mol. Sci. 2021, 22, 8886. [Google Scholar] [CrossRef]
- Wang, W.S.; Lin-Chao, S. Hfq-Antisense RNA I Binding Regulates RNase E-Dependent RNA Stability and ColE1 Plasmid Copy Number. Int. J. Mol. Sci. 2024, 25, 3955. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xi, L.; Qiao, J.; Du, D.; Wang, Y.; Morigen. Involvement of OxyR and Dps in the repression of replication initiation by DsrA small RNA in Escherichia coli. Gene 2023, 882, 147659. [Google Scholar] [CrossRef]
- Li, S.; Edelmann, D.; Berghoff, B.A.; Georg, J.; Evguenieva-Hackenberg, E. Bioinformatic prediction reveals posttranscriptional regulation of the chromosomal replication initiator gene dnaA by the attenuator sRNA rnTrpL in Escherichia coli. RNA Biol. 2021, 18, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Hase, M.; Yoshimi, T.; Ishikawa, Y.; Ohba, A.; Guo, L.; Mima, S.; Makise, M.; Yamaguchi, Y.; Tsuchiya, T.; Mizushima, T. Site-directed mutational analysis for the membrane binding of DnaA protein. Identification of amino acids involved in the functional interaction between DnaA protein and acidic phospholipids. J. Biol. Chem. 1998, 273, 28651–28656. [Google Scholar] [PubMed]
- Hou, Y.; Kumar, P.; Aggarwal, M.; Sarkari, F.; Wolcott, K.M.; Chattoraj, D.K.; Crooke, E.; Saxena, R. The linker domain of the initiator DnaA contributes to its ATP binding and membrane association in E. coli chromosomal replication. Sci. Adv. 2022, 8, eabq6657. [Google Scholar] [CrossRef]
- Mileykovskaya, E.; Dowhan, W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 2009, 1788, 2084–2091. [Google Scholar] [CrossRef]
- Turbant, F.; Machiels, Q.; Waeytens, J.; Wien, F.; Arluison, V. The Amyloid Assembly of the Bacterial Hfq Is Lipid-Driven and Lipid-Specific. Int. J. Mol. Sci. 2024, 25, 1434. [Google Scholar] [CrossRef]
- Ren, J.; Sang, Y.; Lu, J.; Yao, Y.F. Protein Acetylation and Its Role in Bacterial Virulence. Trends Microbiol. 2017, 25, 768–779. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, A.; Tai, C.H.; Chen, J.; Majdalani, N.; Storz, G.; Gottesman, S. An acetyltranferase moonlights as a regulator of the RNA binding repertoire of the RNA chaperone Hfq in Escherichia coli. Proc. Natl. Acad. Sci. USA 2023, 120, e2311509120. [Google Scholar] [CrossRef]
- Ogden, G.B.; Pratt, M.J.; Schaechter, M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell 1988, 54, 127–135. [Google Scholar] [CrossRef]
- Shakibai, N.; Ishidate, K.; Reshetnyak, E.; Gunji, S.; Kohiyama, M.; Rothfield, L. High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB. Proc. Natl. Acad. Sci. USA 1998, 95, 11117–11121. [Google Scholar] [CrossRef]
- Easo George, J.; Basak, R.; Yadav, I.; Tan, C.J.; van Kan, J.A.; Wien, F.; Arluison, V.; van der Maarel, J.R.C. Effect of base methylation on binding and mobility of bacterial protein Hfq on double-stranded DNA. Lab Chip 2024, 24, 5137–5144. [Google Scholar] [CrossRef]
- Wegrzyn, A.; Wrobel, B.; Wegrzyn, G. Altered biological properties of cell membranes in Escherichia coli dnaA and seqA mutants. Mol. Gen. Genet. 1999, 261, 762–769. [Google Scholar] [CrossRef]
- Bloch, S.; Wegrzyn, G.; Arluison, V. The Role of the Hfq Protein in Bacterial Resistance to Antibiotics: A Narrative Review. Microorganisms 2025, 13, 364. [Google Scholar] [CrossRef]
- Firshein, W.; Gelman, I.W. Enrichment of DNA polymerase III activity in a DNA membrane complex purified from Pneumococcus: The possible existence of subcomplexes. Mol. Gen. Genet. 1981, 182, 87–94. [Google Scholar] [CrossRef]
- Sukhodolets, M.V.; Garges, S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 2003, 42, 8022–8034. [Google Scholar] [CrossRef]
- Pool, M.R. Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review). Mol. Membr. Biol. 2005, 22, 3–15. [Google Scholar]
- Bakshi, S.; Choi, H.; Mondal, J.; Weisshaar, J.C. Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes. Mol. Microbiol. 2014, 94, 871–887. [Google Scholar] [CrossRef]
- Bakshi, S.; Siryaporn, A.; Goulian, M.; Weisshaar, J.C. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012, 85, 21–38. [Google Scholar]
- Humphrey, E.D.; Sukhodolets, M.V. Isolation and Partial Characterization of Novel, Structurally Uniform (Hfq(6)) (n>/=8) Assemblies Carrying Accessory Transcription and Translation Factors. Biochemistry 2024, 63, 1647–1662. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Zhang, B.Q.; Chen, Z.Q.; Dong, Y.Q.; You, D.; Zhou, Y.; Ye, B.C. Selective recruitment of stress-responsive mRNAs to ribosomes for translation by acetylated protein S1 during nutrient stress in Escherichia coli. Commun. Biol. 2022, 5, 892. [Google Scholar] [CrossRef]
- Chitra, S.R.; Ramalakshmi, N.; Arunkumar, S.; Manimegalai, P. A Comprehensive Review on DNA Gyrase Inhibitors. Infect. Disord. Drug Targets 2020, 20, 765–777. [Google Scholar] [CrossRef]
- Bartosik, A.A.; Jagura-Burdzy, G. Bacterial chromosome segregation. Acta Biochim. Pol. 2005, 52, 1–34. [Google Scholar] [CrossRef]
- Cayrol, B.; Fortas, E.; Martret, C.; Cech, G.; Kloska, A.; Caulet, S.; Barbet, M.; Trepout, S.; Marco, S.; Taghbalout, A.; et al. Riboregulation of the bacterial actin-homolog MreB by DsrA small noncoding RNA. Integr. Biol. 2015, 7, 128–141. [Google Scholar] [CrossRef]
- Taghbalout, A.; Yang, Q.; Arluison, V. The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J. 2014, 458, 11–22. [Google Scholar] [CrossRef]
- Garvey, N.; St John, A.C.; Witkin, E.M. Evidence for RecA protein association with the cell membrane and for changes in the levels of major outer membrane proteins in SOS-induced Escherichia coli cells. J. Bacteriol. 1985, 163, 870–876. [Google Scholar] [CrossRef]
- Masure, H.R.; Pearce, B.J.; Shio, H.; Spellerberg, B. Membrane targeting of RecA during genetic transformation. Mol. Microbiol. 1998, 27, 845–852. [Google Scholar] [CrossRef]
- Burby, P.E.; Simmons, Z.W.; Schroeder, J.W.; Simmons, L.A. Discovery of a dual protease mechanism that promotes DNA damage checkpoint recovery. PLoS Genet. 2018, 14, e1007512. [Google Scholar] [CrossRef]
- Misra, H.S.; Rajpurohit, Y.S. DNA damage response and cell cycle regulation in bacteria: A twist around the paradigm. Front. Microbiol. 2024, 15, 1389074. [Google Scholar] [CrossRef]
- Rajpurohit, Y.S.; Misra, H.S. Characterization of a DNA damage-inducible membrane protein kinase from Deinococcus radiodurans and its role in bacterial radioresistance and DNA strand break repair. Mol. Microbiol. 2010, 77, 1470–1482. [Google Scholar] [CrossRef]
- Bauermeister, A.; Hahn, C.; Rettberg, P.; Reitz, G.; Moeller, R. Roles of DNA repair and membrane integrity in heat resistance of Deinococcus radiodurans. Arch. Microbiol. 2012, 194, 959–966. [Google Scholar] [CrossRef]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. BioMed Res. Int. 2021, 2021, 1490732. [Google Scholar] [CrossRef]
- Turbant, F.; Waeytens, J.; Blache, A.; Esnouf, E.; Raussens, V.; Wegrzyn, G.; Achouak, W.; Wien, F.; Arluison, V. Interactions and Insertion of Escherichia coli Hfq into Outer Membrane Vesicles as Revealed by Infrared and Orientated Circular Dichroism Spectroscopies. Int. J. Mol. Sci. 2023, 24, 11424. [Google Scholar] [CrossRef]
- Ross, J.A.; Ellis, M.J.; Hossain, S.; Haniford, D.B. Hfq restructures RNA-IN and RNA-OUT and facilitates antisense pairing in the Tn10/IS10 system. RNA 2013, 19, 670–684. [Google Scholar] [CrossRef]
- Ross, J.A.; Wardle, S.J.; Haniford, D.B. Tn10/IS10 transposition is downregulated at the level of transposase expression by the RNA-binding protein Hfq. Mol. Microbiol. 2010, 78, 607–621. [Google Scholar] [CrossRef]
- Ellis, M.J.; Trussler, R.S.; Haniford, D.B. A cis-encoded sRNA, Hfq and mRNA secondary structure act independently to suppress IS200 transposition. Nucleic Acids Res. 2015, 43, 6511–6527. [Google Scholar] [CrossRef]
- Ross, J.A.; Trussler, R.S.; Black, M.D.; McLellan, C.R.; Haniford, D.B. Tn5 transposition in Escherichia coli is repressed by Hfq and activated by over-expression of the small non-coding RNA SgrS. Mob. DNA 2014, 5, 27. [Google Scholar] [CrossRef]
- Ellis, M.J.; Haniford, D.B. Riboregulation of bacterial and archaeal transposition. Wiley Interdiscip. Rev. RNA 2016, 7, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.A.; Hiraga, S.; Ishihama, A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 2000, 5, 613–626. [Google Scholar]
- Azam, T.A.; Ishihama, A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999, 274, 33105–33113. [Google Scholar]
- Macvanin, M.; Edgar, R.; Cui, F.; Trostel, A.; Zhurkin, V.; Adhya, S. Noncoding RNAs binding to the nucleoid protein HU in Escherichia coli. J. Bacteriol. 2012, 194, 6046–6055. [Google Scholar] [CrossRef]
- Qian, Z.; Zhurkin, V.B.; Adhya, S. DNA-RNA interactions are critical for chromosome condensation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 12225–12230. [Google Scholar] [CrossRef]
- Goldberger, O.; Szoke, T.; Nussbaum-Shochat, A.; Amster-Choder, O. Heterotypic phase separation of Hfq is linked to its roles as an RNA chaperone. Cell Rep. 2022, 41, 111881. [Google Scholar] [CrossRef]
- McQuail, J.; Switzer, A.; Burchell, L.; Wigneshweraraj, S. The RNA-binding protein Hfq assembles into foci-like structures in nitrogen starved Escherichia coli. J. Biol. Chem. 2020, 295, 12355–12367. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bloch, S.; Sinden, R.R.; Wien, F.; Węgrzyn, G.; Arluison, V. DNA Transactions in Bacteria and Membranes: A Place for the Hfq Protein? Membranes 2025, 15, 103. https://doi.org/10.3390/membranes15040103
Bloch S, Sinden RR, Wien F, Węgrzyn G, Arluison V. DNA Transactions in Bacteria and Membranes: A Place for the Hfq Protein? Membranes. 2025; 15(4):103. https://doi.org/10.3390/membranes15040103
Chicago/Turabian StyleBloch, Sylwia, Richard R. Sinden, Frank Wien, Grzegorz Węgrzyn, and Véronique Arluison. 2025. "DNA Transactions in Bacteria and Membranes: A Place for the Hfq Protein?" Membranes 15, no. 4: 103. https://doi.org/10.3390/membranes15040103
APA StyleBloch, S., Sinden, R. R., Wien, F., Węgrzyn, G., & Arluison, V. (2025). DNA Transactions in Bacteria and Membranes: A Place for the Hfq Protein? Membranes, 15(4), 103. https://doi.org/10.3390/membranes15040103






