Extracellular miRNAs for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review
Abstract
:1. Introduction
2. Experimental Section
Methods
3. Results
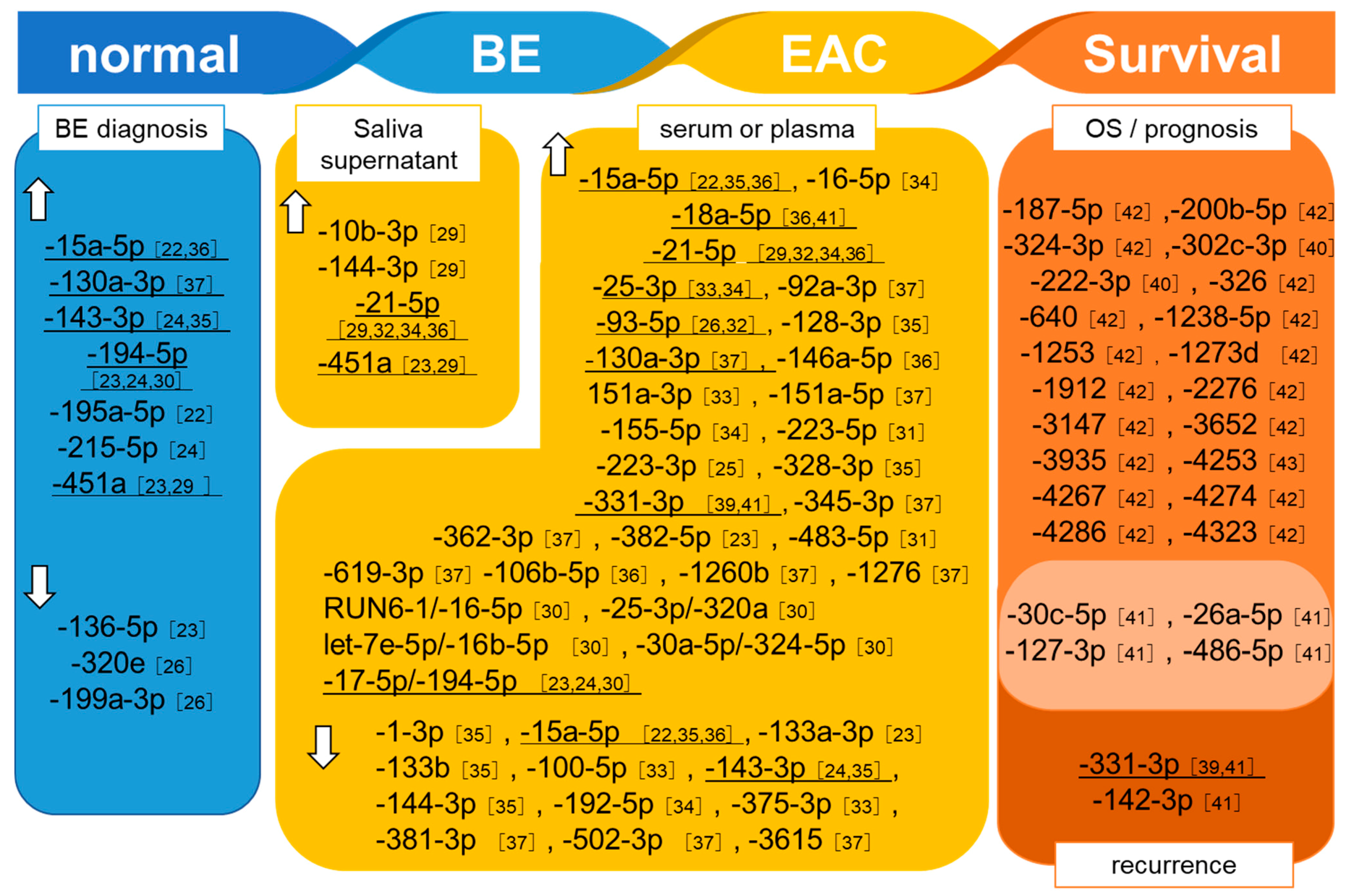
3.1. Detection of BE
| Author, Year [Reference] | Sample Size | Sample | Up-Regulated | Down-Regulated |
|---|---|---|---|---|
| Bansal et al. 2013 [22] | BE (n = 20) GERD (n = 19) | Serum | -15a-5p -195a-5p | |
| Bus et al. 2014 [28] | BE (n = 8) Healthy (n = 6) | Plasma | -194-5p, -451a | -136-5p |
| Cabbi et al. 2016 [24] | BE (n = 8) CLE (n = 12) Esophagitis (n = 10) | Plasma | -143-3p -215-5p -194-5p | |
| Pavlov et al. 2018 [26] | BE (n = 27) SE (n = 19) HGD (n = 17) | Serum | -320e -199a-3p | |
| Wang et al. 2019 [27] | BE (n = 6) Healthy (n = 6) | Serum | -130a-3p |
3.2. Detection of EAC
| Author, Year [Reference] | Sample Size | Sample | Up-Regulated | Down-Regulated |
|---|---|---|---|---|
| Xie et al. 2013 [30] | SCC (n = 32) EAC (n = 4) Healthy (n = 19) | Whole saliva | -10b-3p, -144-3p, -451a | |
| Saliva supernatant | -10b-3p, -144-3p, -21-5p, -451a | |||
| Chiam et al. 2015 [31] | EAC (n = 18) BE (n = 10) Healthy (n = 19) | Serum | RNU6-1/-16-5p, -25-3p/-320a let-7e-5p/-16b-5p, -30a-5p/-324-5p, -17-5p/-194-5p | |
| Warnecke-Eberz et al. 2015 [32] | EAC (n = 59) Healthy (n = 4 | Serum | -223-5p, -483-5p | |
| Bus et al. 2016 [23] | EAC (n = 59) BE (n = 41) Healthy (n = 15) | Plasma | -382-5p | -133a-3p |
| Yan et al. 2016 [33] | Medium serum | -93-5p, -21-5p | ||
| Zhang et al. 2016 [34] | EAC (n = 10) Healthy (n = 11) | Serum | -25-3p, 151a-3p | -100-5p, -375-3p |
| Chen et al. 2017 [35] | EAC (n = 9) Healthy (n = 9) | Serum | -21-5p, -16-5p, -25-3p, -155-5p | -192-5p |
| Fassan et al. 2017 [25] | EAC (n = 30) Dyspeptic patients (n = 20) Atrophic gastritis (n = 15) BE (n = 15) | Plasma | -223-3p | |
| Matsuzaki et al. 2017 [36] | EAC (n = 8) BE (n = 8) Healthy (n = 6) | Plasma | -128-3p, -328-3p | -143-3p, -144-3p, -15a-5p, -1-3p, -133b |
| Miyoshi et al. 2018 [37] | EAC (n = 44) BE (n = 20) Healthy (n = 30) | Serum, | -106b-5p, -93-5p, -146a-5p, -15a-5p, -18a-5p, -21-5p, | |
| Wang et al. 2019 [27] | EAC (n = 40) BE (n = 60) Healthy (n = 30) | Serum | -130a-3p | |
| Fassan et al. 2020 [39] | EAC (n = 8) BE (n = 12) HG-IEN (n = 5) | Serum | -92a-3p, -151a-5p, -362-3p, -345-3p, -619-3p, 1260b, -1276 | -381-3p, -502-3p, -3615 |
3.3. MiRNAs Related to Prognosis and Treatment Responses in EAC Patients
4. Discussion
4.1. BE diagnosis
4.2. EAC Diagnosis
4.3. EAC Recurrence
4.4. Survival and Prognosis
4.5. Potential of Extracellular miRNAs as BE/EAC Diagnosis/Prediction and Prognosis Markers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement.
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xie, S.H.; Lagergren, J. A global assessment of the male predominance in esophageal adenocarcinoma. Oncotarget 2016, 7, 38876–38883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohl, H.; Welch, H.G. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J. Natl. Cancer Inst. 2005, 97, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.G.; Xie, S.H.; Lagergren, J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018, 154, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.B.; Reed, C.E. Epidemiology of esophageal cancer. Surg. Clin. N. Am. 2012, 92, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Laversanne, M.; Brown, L.M.; Devesa, S.S.; Bray, F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: International Trends in Incidence up to 2030. Am. J. Gastroenterol. 2017, 112, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Hanai, M.; Kusano, M.; Kawamura, O.; Shimoyama, Y.; Maeda, M. Epidemiology of Barrett’s esophagus—Comparison of Japan and the West. Nihon Rinsho 2005, 63, 1333–1339. [Google Scholar] [PubMed]
- Nishi, T.; Makuuchi, H.; Ozawa, S.; Shimada, H.; Chino, O. The Present Status and Future of Barrett’s Esophageal Adenocarcinoma in Japan. Digestion 2019, 99, 185–190. [Google Scholar] [CrossRef]
- Clark, G.W. Effect of Helicobacter pylori infection in Barrett’s esophagus and the genesis of esophageal adenocarcinoma. World J. Surg. 2003, 27, 994–998. [Google Scholar] [CrossRef]
- Chow, W.H.; Blaser, M.J.; Blot, W.J.; Gammon, M.D.; Vaughan, T.L.; Risch, H.A.; Perez-Perez, G.I.; Schoenberg, J.B.; Stanford, J.L.; Rotterdam, H.; et al. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 1998, 58, 588–590. [Google Scholar]
- Gavin, A.T.; Francisci, S.; Foschi, R.; Donnelly, D.W.; Lemmens, V.; Brenner, H.; Anderson, L.A. Oesophageal cancer survival in Europe: A EUROCARE-4 study. Cancer Epidemiol. 2012, 36, 505–512. [Google Scholar] [CrossRef]
- Njei, B.; McCarty, T.R.; Birk, J.W. Trends in esophageal cancer survival in United States adults from 1973 to 2009: A SEER database analysis. J. Gastroenterol. Hepatol. 2016, 31, 1141–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Launoy, G.; Bossard, N.; Castro, C.; Manfredi, S. Trends in net survival from esophageal cancer in six European Latin countries: Results from the SUDCAN population-based study. Eur. J. Cancer Prev. 2017, 26, S24–S31. [Google Scholar] [CrossRef] [PubMed]
- Mukaisho, K.I.; Kanai, S.; Kushima, R.; Nakayama, T.; Hattori, T.; Sugihara, H. Barretts’s carcinogenesis. Pathol. Int. 2019, 69, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Abrams, K.R.; De Caestecker, J.S.; Robinson, R.J. Meta analysis: Cancer risk in Barrett’s oesophagus. Aliment. Pharmacol. Ther. 2007, 26, 1465–1477. [Google Scholar] [CrossRef]
- Souza, R.F.; Krishnan, K.; Spechler, S.J. Acid, bile, and CDX: The ABCs of making Barrett’s metaplasia. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G211–G218. [Google Scholar] [CrossRef] [Green Version]
- Savarino, E.; De Bortoli, N.; De Cassan, C.; Della Coletta, M.; Bartolo, O.; Furnari, M.; Ottonello, A.; Marabotto, E.; Bodini, G.; Savarino, V. The natural history of gastro-esophageal reflux disease: A comprehensive review. Dis. Esophagus 2017, 30, 1–9. [Google Scholar] [CrossRef]
- Arnal, M.J.; Arenas, Á.F.; Arbeloa, Á.L. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 2015, 21, 7933–7943. [Google Scholar] [CrossRef]
- Bergman, J.; De Groof, A.J.; Pech, O.; Ragunath, K.; Armstrong, D.; Mostafavi, N.; Lundell, L.; Dent, J.; Vieth, M.; Tytgat, G.N.; et al. An Interactive Web-Based Educational Tool Improves Detection and Delineation of Barrett’s Esophagus-Related Neoplasia. Gastroenterology 2019, 156, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Van der Sommen, F.; Zinger, S.; Curvers, W.L.; Bisschops, R.; Pech, O.; Weusten, B.L.; Bergman, J.J.; De With, P.H.; Schoon, E.J. Computer-aided detection of early neoplastic lesions in Barrett’s esophagus. Endoscopy 2016, 48, 617–624. [Google Scholar] [CrossRef] [Green Version]
- Benaglia, T.; Sharples, L.D.; Fitzgerald, R.C.; Lyratzopoulos, G. Health benefits and cost effectiveness of endoscopic and nonendoscopic cytosponge screening for Barrett’s esophagus. Gastroenterology 2013, 144, 62–73. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Hong, X.; Lee, I.H.; House, J.; Mathur, S.C.; Rastogi, A.; Sharma, P.; Christenson, L.K. Serum exosomal microrna expression can be a novel non-invasive strategy for the screening of barrett’s esophagus. Gastroenterology 2013, 144, S-684. [Google Scholar] [CrossRef]
- Bus, P.; Kestens, C.; Ten Kate, F.J.W.; Peters, W.; Drenth, J.P.H.; Roodhart, J.M.L.; Siersema, P.D.; Van Baal, J.W.P.M. Profiling of circulating microRNAs in patients with Barrett’s esophagus and esophageal adenocarcinoma. J. Gastroenterol. 2016, 51, 560–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabibi, D.; Caruso, S.; Bazan, V.; Castiglia, M.; Bronte, G.; Ingrao, S.; Fanale, D.; Cangemi, A.; Calò, V.; Listì, A.; et al. Analysis of tissue and circulating microRNA expression during metaplastic transformation of the esophagus. Oncotarget 2016, 7, 47821–47830. [Google Scholar] [CrossRef] [Green Version]
- Fassan, M.; Saraggi, D.; Balsamo, L.; Realdon, S.; Scarpa, M.; Castoro, C.; Coati, I.; Salmaso, R.; Farinati, F.; Guzzardo, V.; et al. Early miR-223 Upregulation in Gastroesophageal Carcinogenesis. Am. J. Clin. Pathol. 2017, 147, 301–308. [Google Scholar] [CrossRef] [Green Version]
- Pavlov, K.; Kluiver, J.; Meijer, C.; Boersma-van Ek, W.; Kruyt, F.A.E.; Karrenbeld, A.; Kleibeuker, J.H.; Peters, F.T.M.; Van den Berg, A. Circulating miRNAs in patients with Barrett’s esophagus, high-grade dysplasia and esophageal adenocarcinoma. J. Gastrointest. Oncol. 2018, 9, 1150–1156. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ji, F.; Liu, G.; Wang, W.; Li, Z.; Yue, Y.; Wang, Z. Upregulation of circulating mir130a is correlated with development of barrett’s esophagus and esophageal adenocarcinoma. Onco Targets Ther. 2019, 12, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Van Baal, J.W.; Bus, P.; Kestens, C.; Ten Kate, F.T.; Peters, W.H.; Drenth, J.P.; Roodhart, J.; Voest, E.E.; Siersema, P.D. Comprehensive profiling of plasma microRNAs reveals potential biomarkers for barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology 2014, 146, S-97. [Google Scholar] [CrossRef]
- Song, J.H.; Cheng, Y.; Yang, J.; Wu, W.; Mori, Y.; Olaru, A.V.; Bootwalla, M.; Sharpf, R.; Meltzer, S.J. Circulating miRNAs as noninvasive biomarkers in esophageal adenocarcinoma patients. Cancer Res. 2011, 71, AM2011–AM4960. [Google Scholar]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef]
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnecke-Eberz, U.; Chon, S.H.; Hölscher, A.H.; Drebber, U.; Bollschweiler, E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumor Biol. 2015, 36, 4643–4653. [Google Scholar] [CrossRef] [PubMed]
- Yan, R. Participation of microRNAs in exosome-mediated dysplastic phenotype induction. J. Gastroenterol. Hepatol. 2016, 31, 10. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, X.; Wang, J.; Lopez, J.; Zhou, W.; Yang, L.; Wang, S.E.; Raz, D.J.; Kim, J.Y. Circulating miRNA profile in esophageal adenocarcinoma. Am. J. Cancer Res. 2016, 6, 2713–2721. [Google Scholar] [PubMed]
- Chen, H. Serum exosomal mirnas expression as novel biomarkers for detection of esophageal adenocarcinoma. United Eur. Gastroenterol. J. 2017, 5, A340. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Suzuki, H. Circulating micro RNAs as potential biomarkers to detect transformation of Barrett’s oesophagus to oesophageal Adenocarcinoma. BMJ Open Gastroenterol. 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, J.; Gao, F.; Toden, S.; Shrubsole, M.J.; Scarpa, M.; Whitsett, T.; Inge, L.; Meltzer, S.J.; Teodoro, B.; Coleman, H.G.; et al. A novel non-invasive circulating mirna signature for detection of esophageal adenocarcinoma. Gastroenterology 2018, 154, S-87. [Google Scholar] [CrossRef]
- Craig, M.P.; Rajakaruna, S.; Paliy, O.; Sajjad, M.; Madhavan, S.; Reddy, N.; Zhang, J.; Bottomley, M.; Agrawal, S.; Kadakia, M.P. Differential MicroRNA signatures in the pathogenesis of barrett’s esophagus. Clin. Transl. Gastroenterol. 2020, 11, e00125. [Google Scholar] [CrossRef]
- Fassan, M.; Realdon, S.; Cascione, L.; Hahne, J.C.; Munari, G.; Guzzardo, V.; Arcidiacono, D.; Lampis, A.; Brignola, S.; Dal Santo, L.; et al. Circulating microRNA expression profiling revealed miR-92a-3p as a novel biomarker of Barrett’s carcinogenesis. Pathol. Res. Pract. 2020, 216, 152907. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, J.; Zheng, L.; Ajani, J.A.; Wu, X.; Ye, Y. Serum miR-331-3p predicts tumor recurrence in esophageal adenocarcinoma. Sci. Rep. 2018, 8, 14006. [Google Scholar] [CrossRef]
- Odenthal, M.; Hee, J.; Gockel, I.; Sisic, L.; Schmitz, J.; Stoecklein, N.H.; Driemel, C.; Möhlendick, B.; Schmidt, T.; Knoefel, W.T.; et al. Serum microRNA profiles as prognostic/predictive markers in the multimodality therapy of locally advanced adenocarcinomas of the gastroesophageal junction. Int. J. Cancer 2015, 137, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, X.; Gu, J.; Ye, Y.; Ajani, J.; Wu, X. Circulating miRNAs as potential biomarkers for esophageal adenocarcinoma risk and clinical outcomes. Cancer Res. 2014, 74, AM2014–AM2915. [Google Scholar]
- Zhai, R.; Wei, Y.; Su, L.; Liu, G.; Kulke, M.H.; Wain, J.C.; Christiani, D.C. Whole-miRNome profiling identifies prognostic serum miRNAs in esophageal adenocarcinoma: The influence of Helicobacter pylori infection status. Carcinogenesis 2015, 36, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrick, J.L.; Liao, L.; Wu, X.; Gammon, M.; Vaughan, T.; Cook, M.B. Circulating micrornas in relation to esophageal adenocarcinoma development and survival. Gastroenterology 2018, 154, S-987. [Google Scholar] [CrossRef]
- Ergun, S.; Güney, S.; Temiz, E.; Petrovic, N.; Gunes, S. Significance of miR-15a-5p and CNKSR3 as Novel Prognostic Biomarkers in Non-Small Cell Lung Cancer. Anticancer Agents Med. Chem. 2018, 18, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Kontos, C.K.; Tsiakanikas, P.; Avgeris, M.; Papadopoulos, I.N.; Scorilas, A. miR-15a-5p, A Novel Prognostic Biomarker, Predicting Recurrent Colorectal Adenocarcinoma. Mol. Diagn. Ther. 2017, 21, 453–464. [Google Scholar] [CrossRef]
- Wang, Z.M.; Wan, X.H.; Sang, G.Y.; Zhao, J.D.; Zhu, Q.Y.; Wang, D.M. miR-15a-5p suppresses endometrial cancer cell growth via Wnt/β-catenin signaling pathway by inhibiting WNT3A. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4810–4818. [Google Scholar]
- Zhao, X.Q.; Tang, H.; Yang, J.; Gu, X.Y.; Wang, S.M.; Ding, Y. MicroRNA-15a-5p down-regulation inhibits cervical cancer by targeting TP53INP1 in vitro. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8219–8229. [Google Scholar] [CrossRef]
- Dai, X.; Guo, X.; Liu, J.; Cheng, A.; Peng, X.; Zha, L.; Wang, Z. Circular RNA circGRAMD1B inhibits gastric cancer progression by sponging miR-130a-3p and regulating PTEN and p21 expression. Aging (Albany N. Y.) 2019, 11, 9689–9708. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Shi, H.; Shen, H.; Xu, J.; Zhao, S.; Yao, S.; Jiang, N. MiR-143-3p suppresses the progression of ovarian cancer. Am. J. Transl. Res. 2018, 10, 866–874. [Google Scholar] [PubMed]
- Xia, C.; Yang, Y.; Kong, F.; Kong, Q.; Shan, C. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie 2018, 147, 98–104. [Google Scholar] [CrossRef]
- Xie, F.; Li, C.; Zhang, X.; Peng, W.; Wen, T. MiR-143-3p suppresses tumorigenesis in pancreatic ductal adenocarcinoma by targeting KRAS. Biomed. Pharmacother. 2019, 119, 109424. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Han, B.; Bai, Y.; Ma, X.Y.; Ji, Z.N.; Xiong, Y.; Miao, S.K.; Zhang, Y.Y.; Zhou, L.M. MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer. Cell Death Dis. 2019, 10, 152. [Google Scholar] [CrossRef]
- Li, Z.; Ying, X.; Chen, H.; Ye, P.; Shen, Y.; Pan, W.; Zhang, L. MicroRNA-194 inhibits the epithelial-mesenchymal transition in gastric cancer cells by targeting FoxM1. Dig. Dis. Sci. 2014, 59, 2145–2152. [Google Scholar] [CrossRef]
- Li, T.T.; Gao, X.; Gao, L.; Gan, B.L.; Xie, Z.C.; Zeng, J.J.; Chen, G. Role of upregulated miR-136-5p in lung adenocarcinoma: A study of 1242 samples utilizing bioinformatics analysis. Pathol. Res. Pract. 2018, 214, 750–766. [Google Scholar] [CrossRef]
- Chen, P.; Zhao, L.; Pan, X.; Jin, L.; Lin, C.; Xu, W.; Xu, J.; Guan, X.; Wu, X.; Wang, Y.; et al. Tumor suppressor microRNA-136-5p regulates the cellular function of renal cell carcinoma. Oncol. Lett. 2018, 15, 5995–6002. [Google Scholar] [CrossRef] [Green Version]
- Phatak, P.; Burrows, W.M.; Chesnick, I.E.; Tulapurkar, M.E.; Rao, J.N.; Turner, D.J.; Hamburger, A.W.; Wang, J.Y.; Donahue, J.M. MiR-199a-3p decreases esophageal cancer cell proliferation by targeting p21 activated kinase 4. Oncotarget 2018, 9, 28391–28407. [Google Scholar] [CrossRef]
- Li, L.; Mou, Y.P.; Wang, Y.Y.; Wang, H.J.; Mou, X.Z. miR-199a-3p targets ETNK1 to promote invasion and migration in gastric cancer cells and is associated with poor prognosis. Pathol. Res. Pract. 2019, 215, 152511. [Google Scholar] [CrossRef]
- Fassan, M.; Volinia, S.; Palatini, J.; Pizzi, M.; Fernandez-Cymering, C.; Balistreri, M.; Realdon, S.; Battaglia, G.; Souza, R.; Odze, R.D.; et al. MicroRNA Expression Profiling in the Histological Subtypes of Barrett’s Metaplasia. Clin. Transl. Gastroenterol. 2013, 4, e34. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Merhautova, J.; Machackova, T.; Gutierrez-Garcia, I.; Garcia-Solano, J.; Radova, L.; Brchnelova, D.; Slaba, K.; Svoboda, M.; Halamkova, J.; et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis 2017, 6, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.B.; Zhu, M.N.; Zhu, X.L. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio 2019, 9, 1957–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drahos, J.; Schwameis, K.; Orzolek, L.D.; Hao, H.; Birner, P.; Taylor, P.R.; Pfeiffer, R.M.; Schoppmann, S.F.; Cook, M.B. MicroRNA Profiles of Barrett’s Esophagus and Esophageal Adenocarcinoma: Differences in Glandular Non-native Epithelium. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Cui, R.; Bahr, J.; Zanesi, N.; Luo, Z.; Meng, W.; Liang, G.; Croce, C.M. miR-130a Deregulates PTEN and Stimulates Tumor Growth. Cancer Res. 2017, 77, 6168–6178. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.Y.; Cui, J.Y.; Yuan, J.; Wang, X. MiR-451a suppressed cell migration and invasion in non-small cell lung cancer through targeting ATF2. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5554–5561. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, H.; Yang, X.; Ge, Y.; Zhang, C.; Qin, Q.; Lu, J.; Zhan, L.; Cheng, H.; Sun, X. MicroRNA-21 is a novel promising target in cancer radiation therapy. Tumour Biol. 2014, 35, 3975–3979. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, X.; Tian, W.; Yin, X.; Wang, J.; Yang, H. The role of TGF-β1-miR-21-ROS pathway in bystander responses induced by irradiated non-small-cell lung cancer cells. Br. J. Cancer 2014, 111, 772–780. [Google Scholar] [CrossRef]
- Chen, J.; Gu, Y.; Shen, W. MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4566–4576. [Google Scholar]
- Masoudi, M.S.; Mehrabian, E.; Mirzaei, H. MiR-21: A key player in glioblastoma pathogenesis. J. Cell. Biochem. 2018, 119, 1285–1290. [Google Scholar] [CrossRef]
- Yang, C.H.; Pfeffer, S.R.; Sims, M.; Yue, J.; Wang, Y.; Linga, V.G.; Paulus, E.; Davidoff, A.M.; Pfeffer, L.M. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J. Biol. Chem. 2015, 290, 6037–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.Y.; Wang, L.M.; Liu, J.; Huang, X.; Liu, J.; Zhang, Y.F. MiR-21 Suppresses Anoikis through Targeting PDCD4 and PTEN in Human Esophageal Adenocarcinoma. Curr. Med. Sci. 2018, 38, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Winther, M.; Alsner, J.; Tramm, T.; Baeksgaard, L.; Holtved, E.; Nordsmark, M. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol. 2015, 54, 1582–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Chen, Z.; Zhao, X.; Wang, J.; Ding, D.; Wang, Z.; Tan, F.; Tan, X.; Zhou, F.; Sun, J.; et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2012, 421, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Z.; Cheng, W.C.; Chen, S.F.; Nieh, S.; O’Connor, C.; Liu, C.L.; Tsai, W.W.; Wu, C.J.; Martin, L.; Lin, Y.S.; et al. miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat. Cell Biol. 2017, 19, 1286–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.Y.; Han, X.M.; Zhao, X.L.; Cheng, X.M.; Zhang, Y. MiR-93-5p promotes cervical cancer progression by targeting THBS2/MMPS signal pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5113–5121. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Tsugawa, H.; Masaoka, T.; Mori, H.; Watanabe, M.; Kanai, T.; Suzuki, H. Adiponectin ameliorates carcinogenic pathway depending on miR-221/222 in esophageal adenocarcinoma cells. Gastroenterology 2015, 148, S347. [Google Scholar] [CrossRef]
- Chava, S.; Reynolds, C.P.; Pathania, A.S.; Gorantla, S.; Poluektova, L.Y.; Coulter, D.W.; Gupta, S.C.; Pandey, M.K.; Challagundla, K.B. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol. Oncol. 2020, 14, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Yamada, Y.; Arai, T.; Okato, A.; Idichi, T.; Kato, M.; Koshizuka, K.; Ichikawa, T.; Seki, N. Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: Targeted genes are involved in bladder cancer pathogenesis. J. Hum. Genet. 2018, 63, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Matsui, D.; Zaidi, A.H.; Martin, S.A.; Omstead, A.N.; Kosovec, J.E.; Huleihel, L.; Saldin, L.T.; DiCarlo, C.; Silverman, J.F.; Hoppo, T.; et al. Primary tumor microRNA signature predicts recurrence and survival in patients with locally advanced esophageal adenocarcinoma. Oncotarget 2016, 7, 81281–81291. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ma, S.P.; Yang, D.; Liu, Y.; Wang, Y.P.; Lin, T.; Li, Y.X.; Yang, S.H.; Zhang, W.C.; Wang, X.L. miR-142-3p Suppresses Cell Growth by Targeting CDK4 in Colorectal Cancer. Cell. Physiol. Biochem. 2018, 51, 1969–1981. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Ghasabi, M.; Shirjang, S.; Dehghan, R.; Montazeri, V.; Holmskov, U.; Kazemi, T.; Duijf, P.; Gjerstorff, M.; et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J. Cell. Physiol. 2019, 234, 9816–9825. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, H.; Li, X.; Tian, X.; Peng, B.; Liu, S.; Zhan, T.; Wan, Y.; Chen, W.; Li, Y.; et al. miR-331-3p functions as an oncogene by targeting ST7L in pancreatic cancer. Carcinogenesis 2018, 39, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, X.; Wang, Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J. Gynecol. Oncol. 2018, 29, e95. [Google Scholar] [CrossRef]
- Zhao, D.; Sui, Y.; Zheng, X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol. Rep. 2016, 35, 1075–1082. [Google Scholar] [CrossRef] [Green Version]
- Buranjiang, G.; Kuerban, R.; Abuduwanke, A.; Li, X.; Kuerban, G. MicroRNA-331-3p inhibits proliferation and metastasis of ovarian cancer by targeting RCC2. Arch. Med. Sci. 2019, 15, 1520–1529. [Google Scholar] [CrossRef]
- Guo, X.; Guo, L.; Ji, J.; Zhang, J.; Zhang, J.; Chen, X.; Cai, Q.; Li, J.; Gu, Q.; Liu, B.; et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 2010, 398, 1–6. [Google Scholar] [CrossRef]
- Fukumoto, I.; Hanazawa, T.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Goto, Y.; Nishikawa, R.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer 2015, 112, 891–900. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.M.; Li, G.Z.; Han, M.; Xu, H.L.; Huang, K.M. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed. Pharmacother. 2017, 93, 554–560. [Google Scholar] [CrossRef]
- Song, S.; Long, M.; Yu, G.; Cheng, Y.; Yang, Q.; Liu, J.; Wang, Y.; Sheng, J.; Wang, L.; Wang, Z.; et al. Urinary exosome miR-30c-5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5. J. Cell. Mol. Med. 2019, 23, 6755–6765. [Google Scholar] [CrossRef]
- Ji, L.; Zhu, Z.N.; He, C.J.; Shen, X. MiR-127-3p targets KIF3B to inhibit the development of oral squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, L.; Wu, H.; Wang, K.; Gu, D. MiR-127-3p inhibits cell growth and invasiveness by targeting ITGA6 in human osteosarcoma. IUBMB Life 2018, 70, 411–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, L.; Yang, Q.; Yuan, J.; Miao, Q.; Duan, L.; Li, F.; Wang, S. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol. Rep. 2016, 36, 2563–2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.C.; Yu, Y.; Hou, L.K.; Sun, X.H.; Ge, J.; Zhang, B.; Wang, X. miR-142-3p inhibits cancer cell proliferation by targeting CDC25C. Cell Prolif. 2016, 49, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Ménard, S.; Pupa, S.M.; Campiglio, M.; Tagliabue, E. Biologic and therapeutic role of HER2 in cancer. Oncogene 2003, 22, 6570–6578. [Google Scholar] [CrossRef] [Green Version]
- Yuan, R.; Zhang, X.; Fang, Y.; Nie, Y.; Cai, S.; Chen, Y.; Mo, D. mir-127-3p inhibits the proliferation of myocytes by targeting KMT5a. Biochem. Biophys. Res. Commun. 2018, 503, 970–976. [Google Scholar] [CrossRef]
- Li, C.; Zheng, X.; Li, W.; Bai, F.; Lyu, J.; Meng, Q.H. Serum miR-486-5p as a diagnostic marker in cervical cancer: With investigation of potential mechanisms. BMC Cancer 2018, 18, 61. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.J.; Yuan, W.D.; Yuan, J.Q.; Yuan, K.; Wang, Y. miR-486-5p functions as an oncogene by targeting PTEN in non-small cell lung cancer. Pathol. Res. Pract. 2018, 214, 700–705. [Google Scholar] [CrossRef]
- Tian, F.; Wang, J.; Ouyang, T.; Lu, N.; Lu, J.; Shen, Y.; Bai, Y.; Xie, X.; Ge, Q. MiR-486-5p Serves as a Good Biomarker in Nonsmall Cell Lung Cancer and Suppresses Cell Growth With the Involvement of a Target PIK3R1. Front. Genet. 2019, 10, 688. [Google Scholar] [CrossRef] [Green Version]
- Noto, J.M.; Peek, R.M. The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front. Cell. Infect. Microbiol. 2011, 1, 21. [Google Scholar] [CrossRef] [Green Version]
- Matsushima, K.; Isomoto, H.; Inoue, N.; Nakayama, T.; Hayashi, T.; Nakayama, M.; Nakao, K.; Hirayama, T.; Kohno, S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer 2011, 128, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, J.; Zou, Y.; Jiao, Y.; Huang, Y.; Fan, L.; Li, X.; Yu, H.; He, C.; Wei, W.; et al. MicroRNA-143-3p, up-regulated in H. pylori-positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget 2017, 8, 28711–28724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, A.P.; Badr, A.S.; Topalovski, M.; Cherian, R.; Dixon, A.; Hassanein, R.S. Prospective evaluation of the prevalence of gastric Helicobacter pylori infection in patients with GERD, Barrett’s esophagus, Barrett’s dysplasia, and Barrett’s adenocarcinoma. Am. J. Gastroenterol. 2000, 95, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, J.H.; Shaheen, N.J. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology 2015, 149, 302–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egyud, M.; Tejani, M.; Pennathur, A.; Luketich, J.; Sridhar, P.; Yamada, E.; Ståhlberg, A.; Filges, S.; Krzyzanowski, P.; Jackson, J.; et al. Detection of Circulating Tumor DNA in Plasma: A Potential Biomarker for Esophageal Adenocarcinoma. Ann. Thorac. Surg. 2019, 108, 343–349. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Aspects Med. 2020, 72, 100825. [Google Scholar] [CrossRef]
- Buschmann, D.; Haberberger, A.; Kirchner, B.; Spornraft, M.; Riedmaier, I.; Schelling, G.; Pfaffl, M.W. Toward reliable biomarker signatures in the age of liquid biopsies—How to standardize the small RNA-Seq workflow. Nucleic Acids Res. 2016, 44, 5995–6018. [Google Scholar] [CrossRef]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef]

| miRNA | Functional Role | Target | Cancer Type | Sample |
|---|---|---|---|---|
| miR-15-a-5p | Tumor promoter Tumor suppressor | TP53INP1 [48] WNT3A [47] | Cervical cancer Endometrial Cancer | 30 tissue 8 tissue |
| miR-130a-3p | Inhibit cell proliferation, migration, and invasion | RAB5B [50] PTEN, p21 [64] | Breast cancer Gastric cancer | 40 tissue 30 tissue |
| miR-143-3p | Tumor suppressor | TAK1 [51] MAPK7 [52] KRAS [53] | Ovarian cancer Breast cancer PDAC | 4 cell lines 37 tissue |
| miR-215-5p | Tumor suppressor | EGFR [61] Sox9 [62] | Colorectal cancer Breast cancer | 252 tissue 39 tissue |
| miR-194-5p | Inhibiting cell migration and invasion during cancer progression | FoxM1 [55] | Gastric cancer | 50 tissue |
| miR-451a | Inhibits the migratory and invasive abilities | ATF2 [65] | NSCLC | 55 tissue |
| miR-199a-3p | Prognostic indicator Tumor suppressor | ETNK1 [59] PAK4 [58] | Gastric cancer Esophageal cancer | 39 tissue |
| miRNA | Functional Role | Target | Cancer Type | Sample |
|---|---|---|---|---|
| miR-21-5p | Promotes anoikis resistance and metastasis | TGFβ1 [68] TIMP3 [69] IGFBP3 [67] FBXO11 [71] | NSCLC RCC GBM | 104 tissue |
| miR-25-3p | Promote migration invasion | CDH1 [74] | SCC | |
| miR-93-5p | Promote progression | PTEN [75] THBS2/MMPS [76] | Breast cancer Cervical cancer | 28 tissue |
| miR-15a-5p | Tumor suppressor Promotor | E2F1, MYCN [78] TP53INP1 [48] | Neuroblastoma Cervical cancer | 30 tissue |
| miRNA | Functional Role | Target | Cancer Type | Sample |
|---|---|---|---|---|
| miR-331-3p | Tumor suppressor | HER2 [84,85] RCC2 [86] E2F1 [87] | Pancreatic cancer Colorectal cancer Cervical cancer Ovarian cancer Gastric cancer | 44 plasma 29 tissue 20 tissue |
| miR-26a-5p | Tumor suppressor | TMEM184B [88] | OSCC | 36 tissue |
| miR-30c-5p | Tumor suppressor | MTA1 [89] HSPA5 [90] | Gastric cancer RCC, Bladder cancer, Prostate cancer | 40 tissue 130 urine |
| miR-127-3p | Tumor suppressor | KIF3B [91] ITGA6 [92] BAG5 [93] | OSCC Osteosarcoma Ovarian cancer | 45 tissue 20 tissue |
| miR-142-3p | Tumor suppressor | CDK4 [81] CDC25C [94] Beach-1 [82] | Colorectal Cancer Breast Cancer | 116 tissue 42 tissue |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inokuchi, K.; Ochiya, T.; Matsuzaki, J. Extracellular miRNAs for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review. J. Clin. Med. 2021, 10, 117. https://doi.org/10.3390/jcm10010117
Inokuchi K, Ochiya T, Matsuzaki J. Extracellular miRNAs for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review. Journal of Clinical Medicine. 2021; 10(1):117. https://doi.org/10.3390/jcm10010117
Chicago/Turabian StyleInokuchi, Kazumi, Takahiro Ochiya, and Juntaro Matsuzaki. 2021. "Extracellular miRNAs for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review" Journal of Clinical Medicine 10, no. 1: 117. https://doi.org/10.3390/jcm10010117
APA StyleInokuchi, K., Ochiya, T., & Matsuzaki, J. (2021). Extracellular miRNAs for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review. Journal of Clinical Medicine, 10(1), 117. https://doi.org/10.3390/jcm10010117





