Gut Bacterial Dysbiosis in Children with Intractable Epilepsy
Abstract
1. Introduction
2. Experimental Section
2.1. Study Design
2.2. 16S rRNA Gene Sequencing
2.3. Taxonomic Comparison and Diversity Indices
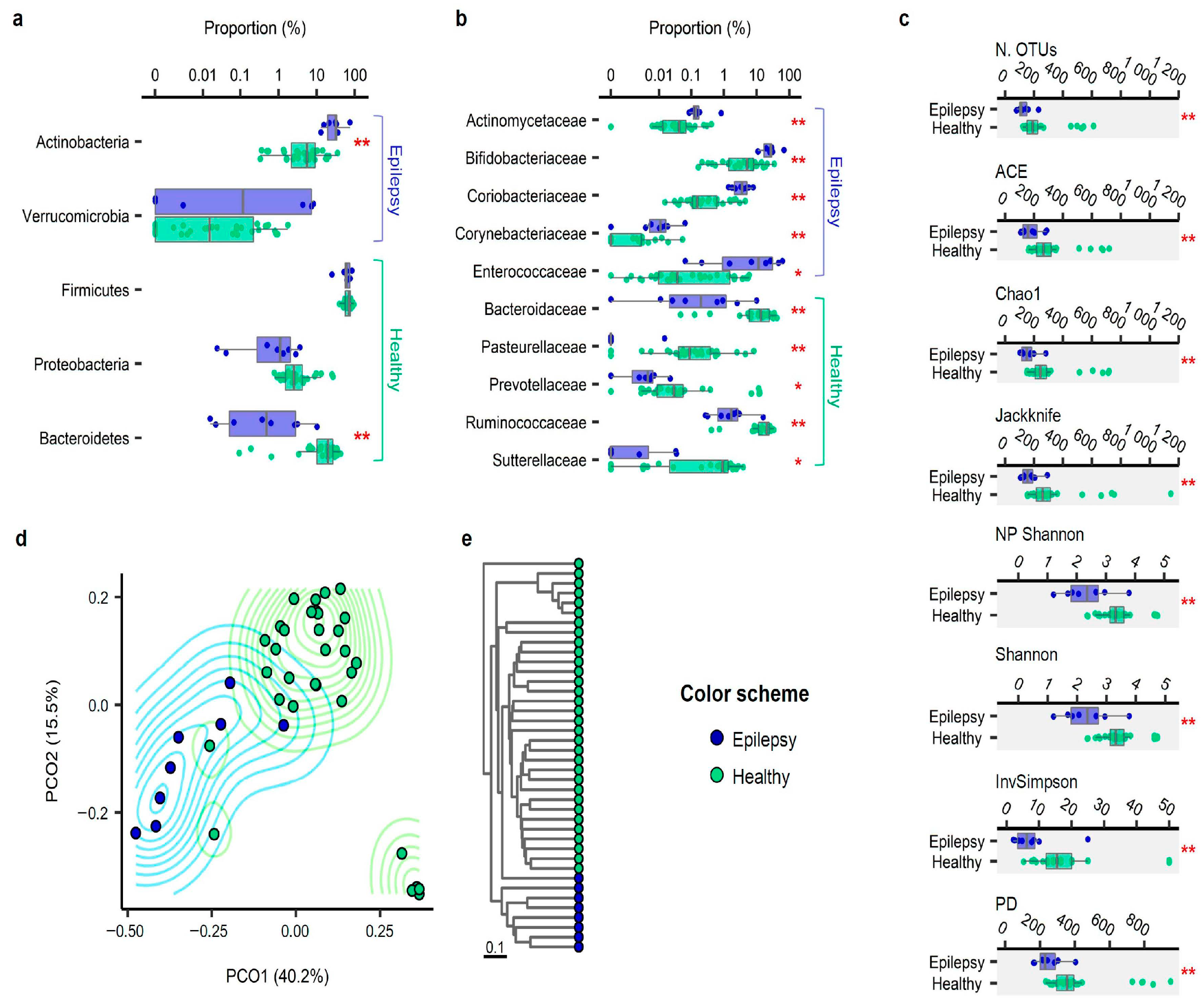
3. Results
3.1. Study Population
3.2. Taxonomic Compositions
3.3. Alpha Diversity
3.4. Beta Diversity
3.5. Effect of Constipation and a Liquid- or Formula-Based Diet
3.6. Taxonomic and Functional Biomarker Discovery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E.; Centers, T.U.N.O.P.M. Gut microbiota in early pediatric multiple sclerosis: A case−control study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, C.B.; Shannon, K.M.; Kordower, J.H.; Voigt, R.M.; Shaikh, M.; Jaglin, J.A.; Estes, J.D.; Dodiya, H.B.; Keshavarzian, A. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson’s disease. PLoS ONE 2011, 6, e28032. [Google Scholar] [CrossRef]
- Hilton, D.A.; Stephens, M.; Kirk, L.; Edwards, P.; Potter, R.; Zajicek, J.; Broughton, E.; Hagan, H.; Carroll, C. Accumulation of α-synuclein in the bowel of patients in the pre-clinical phase of Parkinson’s disease. Acta Neuropathol. 2013, 127, 235–241. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C.; et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- GBD 2016 Epilepsy Collaborators. Global, regional, and national burden of epilepsy, 1990–2016: A systematic analysis for the Global Burden of Disease Study. Lancet Neurol. 2019, 18, 357–375. [Google Scholar] [CrossRef]
- Chen, T.; Giri, M.; Xia, Z.; Subedi, Y.N.; Li, Y. Genetic and epigenetic mechanisms of epilepsy: A review. Neuropsychiatr. Dis. Treat. 2017, 13, 1841–1859. [Google Scholar] [CrossRef]
- Partemi, S.; Vidal, M.C.; Striano, P.; Campuzano, O.; Allegue, C.; Pezzella, M.; Elia, M.; Parisi, P.; Belcastro, V.; Casellato, S.; et al. Genetic and forensic implications in epilepsy and cardiac arrhythmias: A case series. Int. J. Leg. Med. 2014, 129, 495–504. [Google Scholar] [CrossRef]
- Coll, M.; Allegue, C.; Partemi, S.; Mates, J.; Del Olmo, B.; Campuzano, O.; Pascali, V.; Lglesias, A.; Striano, P.; Olivia, A.; et al. Genetic investigation of sudden unexpected death in epilepsy cohort by panel target resequencing. Int. J. Legal Med. 2016, 130, 331–339. [Google Scholar] [CrossRef]
- Dahlin, M.; Prast-Nielsen, S. The gut microbiome and epilepsy. EBioMedicine 2019, 44, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Sutter, R.; Rüegg, S.; Tschudin-Sutter, S. Seizures as adverse events of antibiotic drugs: A systematic review. Neurology 2015, 85, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Cui, B.-T.; Zhang, T.; Li, P.; Long, C.-Y.; Ji, G.-Z.; Zhang, F.-M. Fecal microbiota transplantation cured epilepsy in a case with Crohn’s disease: The first report. World J. Gastroenterol. 2017, 23, 3565–3568. [Google Scholar] [CrossRef]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.J.; Hsiao, E.Y. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 2018, 173, 1728–1741.e1713. [Google Scholar] [CrossRef]
- Lindefeldt, M.; Eng, A.; Darban, H.; Bjerkner, A.; Zetterström, C.K.; Allander, T.; Andersson, B.; Borenstein, E.; Dahlin, M.; Prast-Nielsen, S. The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. NPJ Biofilms Microbiomes 2019, 5, 1–13. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.; Zhou, Y.; Yu, L.; Zhang, L.; Wang, Y. Altered gut microbiome composition in children with refractory epilepsy after ketogenic diet. Epilepsy Res. 2018, 145, 163–168. [Google Scholar] [CrossRef]
- Peng, A.; Qiu, X.; Lai, W.; Zhang, L.; Zhu, X.; He, S.; Duan, J.; Chen, L. Altered composition of the gut microbiome in patients with drug-resistant epilepsy. Epilepsy Res. 2018, 147, 102–107. [Google Scholar] [CrossRef]
- Xie, G.; Zhou, Q.; Qiu, C.-Z.; Dai, W.-K.; Wang, H.-P.; Li, Y.-H.; Liao, J.-X.; Lu, X.-G.; Lin, S.-F.; Ye, J.-H.; et al. Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J. Gastroenterol. 2017, 23, 6164–6171. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimaraes, V.D.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, K.; Meyer, K.M.; Aagaard, K.M.; Wilmes, P. The human gut microbiome in health: Establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 2016, 18, 2103–2116. [Google Scholar] [CrossRef]
- Stanislawski, M.; Dabelea, D.; Lange, L.A.; Wagner, B.D.; Lozupone, C. Gut microbiota phenotypes of obesity. NPJ Biofilms Microbiomes 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Lee, S.A.; Lim, J.Y.; Kim, B.-S.; Cho, S.J.; Kim, N.Y.; Bin Kim, O.; Kim, Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr. Res. Pract. 2015, 9, 242–248. [Google Scholar] [CrossRef]
- Murtaza, N.; Burke, L.M.; Vlahovich, N.; Charlesson, B.; Neill, H.O.; Ross, M.L.; Campbell, K.L.; Krause, L.; Morrison, M. The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers. Nutrients 2019, 11, 261. [Google Scholar] [CrossRef]
- Khine, W.W.T.; Zhang, Y.; GoIe, G.J.Y.; Wong, M.S.; Liong, M.; Lee, Y.Y.; Cao, H.; Lee, Y.-K. Gut microbiome of pre-adolescent children of two ethnicities residing in three distant cities. Sci. Rep. 2019, 9, 7831. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Reddel, S.; Putignani, L.; Del Chierico, F. The Impact of Low-FODMAPs, Gluten-Free, and Ketogenic Diets on Gut Microbiota Modulation in Pathological Conditions. Nutrients 2019, 11, 373. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, Å.; Edlund, C.; Nord, C.E. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001, 1, 101–114. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nat. Cell Biol. 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björkstén, B.; Engstrand, L.; Andersson, A.F.; Filion, K.B.; et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hämäläinen, A.-M.; Härkönen, T.; Ryhänen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016, 8, 343ra381. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Haiser, H.J.; Gootenberg, D.B.; Chatman, K.; Sirasani, G.; Balskus, E.P.; Turnbaugh, P. Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthella lenta. Science 2013, 341, 295–298. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Tang, F.; Hartz, A.M.S.; Bauer, B. Drug-Resistant Epilepsy: Multiple Hypotheses, Few Answers. Front. Neurol. 2017, 8, 301. [Google Scholar] [CrossRef]
- Orelle, C.; Mathieu, K.; Jault, J.-M. Multidrug ABC transporters in bacteria. Res. Microbiol. 2019, 170, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The Microbiome and Host Behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M. Microbial Endocrinology and the Microbiota-Gut-Brain Axis. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLCL: Heidelberg, Germany, 2014; Volume 817, pp. 3–24. [Google Scholar]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- Stokes, J.M.; Davis, J.H.; Mangat, C.S.; Williamson, J.R.; Brown, E.D. Discovery of a small molecule that inhibits bacterial ribosome biogenesis. eLife 2014, 3, e03574. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 2019, 363, eaat9931. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
| Study | Population 1 | Patients’ Age (Years) 2 | Excluded or Matched Variables | Profile of Intractable Epilepsy Compared to the Controls 1 | Change Before and After the Ketogenic Diet 1 | |||
|---|---|---|---|---|---|---|---|---|
| Taxon | α- Diversity | β- Diversity | Biomarker | |||||
| Our study | IE (n = 8), HC (n = 32) | 3.17 (1.16–6.92) | Age, obesity, delivery methods, diet, antibiotic exposure | ↓ Bacteroidetes, Proteobacteria ↑ Actinobacteria | Lower | SP | ↑ E.faecium, B.longum, E.lenta ↑ ABCT | NA |
| Lindefeldt et al. [16] | IE (n = 12), Healthy parents (n = 11) | 7.7 (2.2–15.3) | NA | NA | Lower | NA | ↑ Hemin Transport system ↓ Carbohydrate metabolism | ↑ E. coli ↓ Bifidobacteria, E. rectale, Dialister α-diversity: NS |
| Zhang et al. [17] | IE (n = 20) | 4.2 (1.2–10.3) | Obesity, antibiotic exposure | NA | NA | NA | NA | ↑ Bacteroides ↓ Firmicutes, Actinobacteria α-diversity: NS, β-diversity: SP |
| Peng et al. [18] | IE (n = 42), HC (n = 65), drug responsive epilepsy (n = 49) | 28.4 (5–50) | Antibiotic exposure | ↓ Bacteroidetes ↑ Firmicutes, rare species | Higher | SP | ↑ ABCT ↓ Glucose and lipid metabolism | NA |
| Xie et al. [19] | IE (n = 14), HC (n = 30) | 1.95 (0.8–3.3) | Antibiotic exposure | ↓ Bacteroidetes ↑ Firmicutes | Lower | SP | NA | ↑ Bacteroidetes ↓ Proteobacteria |
| No. | Age (Years) | Sex | Epilepsy Duration (Years) | Epilepsy Type | Etiology | Seizure Frequency | Past AEDs | Current AEDs | Type of Food |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.15 | M | 0.65 | Infantile spasms | Unknown | >10 clusters/d | - | VGB, CLB, LVT, TPM | Artificial formula (oral) |
| 2 | 4.81 | F | 4.31 | Generalized | Unknown | 3/d | - | VPA, LVT | Solid |
| 3 | 2.63 | F | 2.05 | Focal and generalized | Genetic | 1/mo | LVT | VPA, CLB | Solid |
| 4 | 4.52 | F | 3.44 | Focal | Unknown | 15/d | - | OXC, LVT | Solid |
| 5 | 6.92 | F | 6.26 | Focal and generalized | Unknown | 3/d | VPA | VPA, PB, CLB, LVT, TPM, LTG | Enteral formula (tube feeding) |
| 6 | 3.52 | M | 2.93 | Generalized | Genetic | 25/d | - | VPA, OXC, LTG, TPM | Solid |
| 7 | 2.82 | F | 2.40 | Infantile spasms | Structural | 3–4 clusters/d | VGB, TPM | VPA, OXC | Solid |
| 8 | 1.58 | M | 1.16 | Infantile spasms | Structural | 5–6 clusters/d | TPM | VGB, OXC | Solid |
| Group | Epilepsy (n = 8) | Control (n = 32) | p |
|---|---|---|---|
| Sex 1 | 0.812 | ||
| Female | 5 (62.5) | 16 (50.0) | |
| Male | 3 (37.5) | 16 (50.0) | |
| Age (years) 2 | 3.17 (2.11; 4.67) | 3.67 (1.33; 5.27) | 0.946 |
| Body mass index 3 | 16.6 ± 2.3 | 16.4 ± 1.8 | 0.799 |
| Mode of delivery 1 | 0.804 | ||
| Vaginal | 5 (62.5) | 13 (76.5) | |
| Cesarean | 3 (37.5) | 4 (23.5) | |
| Liquid diet 1 | 2 (25.0), on a liquid diet 4 | 0 | 0.174 |
| Constipation 1 | 3 (37.5) | 5 (31.3) | 1.000 |
| Marker Type | Epilepsy Biomarkers | Healthy Biomarkers | ||
|---|---|---|---|---|
| Names | N | Names | N | |
| Taxonomic 1 | 17 | 18 | ||
| Species | B. longum group, E. lenta, E. faecium group | 3 | Bacteroides vulgatus, Faecalibacterium prausnitzii group | 2 |
| Genus | Bifidobacterium, Eggerthella, Enterococcus | 3 | Bacteroides, Faecalibacterium, Lachnospira, Roseburia, Veillonella | 5 |
| Family | Bifidobacteriaceae, Coriobacteriaceae, Enterococcaceae, Streptococcaceae | 4 | Bacteroidaceae, Ruminococcaceae, Veillonellaceae | 3 |
| Order | Bifidobacteriales, Coriobacteriales, Lactobacillales | 3 | Bacteroidales, Clostridiales, Veillonellales | 3 |
| Class | Actinobacteria_c, Bacilli, Coriobacteriia | 3 | Bacteroidia, Clostridia, Negativicutes | 3 |
| Phylum | Actinobacteria | 1 | Bacteroidetes, Proteobacteria | 2 |
| Functional 2 | 11 | 1 | ||
| Module | Putative multiple sugar transport system (M00207), peptides/nickel transport system (M00239), energy-coupling factor transport system (M00582), putative ABC transport system (M00258), ABC-2 type transport system (M00254), PTS system, cellobiose-specific II component (M00275), PTS system, beta-glucoside-specific II component (M00271), putative aldouronate transport system (M00603) | 8 | Cobalamin biosynthesis, cobinamide to cobalamin (M00122) | 1 |
| Pathway | ABC transporters (ko02010), quorum sensing (ko02024), starch and sucrose metabolism (ko00500) | 3 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Kim, N.; Shim, J.O.; Kim, G.-H. Gut Bacterial Dysbiosis in Children with Intractable Epilepsy. J. Clin. Med. 2021, 10, 5. https://doi.org/10.3390/jcm10010005
Lee K, Kim N, Shim JO, Kim G-H. Gut Bacterial Dysbiosis in Children with Intractable Epilepsy. Journal of Clinical Medicine. 2021; 10(1):5. https://doi.org/10.3390/jcm10010005
Chicago/Turabian StyleLee, Kihyun, Namil Kim, Jung Ok Shim, and Gun-Ha Kim. 2021. "Gut Bacterial Dysbiosis in Children with Intractable Epilepsy" Journal of Clinical Medicine 10, no. 1: 5. https://doi.org/10.3390/jcm10010005
APA StyleLee, K., Kim, N., Shim, J. O., & Kim, G.-H. (2021). Gut Bacterial Dysbiosis in Children with Intractable Epilepsy. Journal of Clinical Medicine, 10(1), 5. https://doi.org/10.3390/jcm10010005





