Prognosis in Myelodysplastic Syndromes: The Clinical Challenge of Genomic Integration
Abstract
1. Introduction
1.1. Past and Present MDS Prognostic Models
1.2. The Classical System and Its Modifications
1.3. Revised System
2. Incoming MDS Prognostic Models
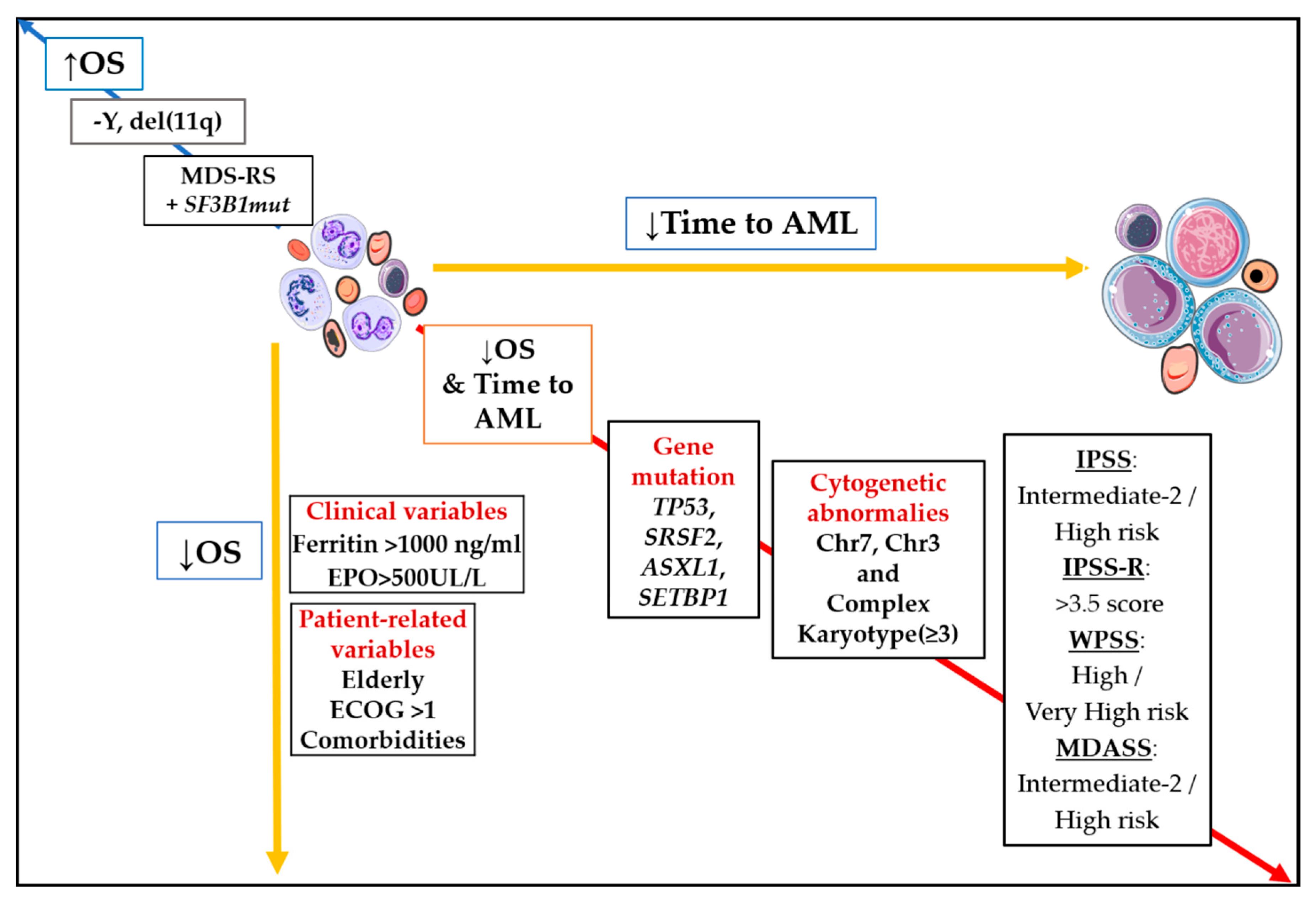
New Approaches: Machine Learning, Big Data, and “Omics” Integration
3. Conclusions and Concerns
Funding
Conflicts of Interest
References
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Shreve, J.; Nazha, A. Novel Prognostic Models for Myelodysplastic Syndromes. Hematol. Oncol. Clin. N. Am. 2020, 34, 369–378. [Google Scholar] [CrossRef]
- Kantarjian, H.; O’Brien, S.; Ravandi, F.; Cortes, J.; Shan, J.; Bennett, J.M.; List, A.; Fenaux, P.; Sanz, G.; Issa, J.-P.; et al. Proposal for a new risk model in myelodysplastic syndrome that accounts for events not considered in the original International Prognostic Scoring System. Cancer 2008, 113, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Schanz, J.; Tüchler, H.; Solé, F.; Mallo, M.; Luño, E.; Cervera, J.; Granada, I.; Hildebrandt, B.; Slovak, M.L.; Ohyashiki, K.; et al. New Comprehensive Cytogenetic Scoring System for Primary Myelodysplastic Syndromes (MDS) and Oligoblastic Acute Myeloid Leukemia after MDS Derived from an International Database Merge. J. Clin. Oncol. 2012, 30, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Germing, U.; Kuendgen, A.; Della Porta, M.G.; Pascutto, C.; Invernizzi, R.; Giagounidis, A.; Hildebrandt, B.; Bernasconi, P.; Knipp, S.; et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J. Clin. Oncol. 2007, 25, 3503–3510. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Della Porta, M.G.; Tuechler, H.; Malcovati, L.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; et al. Validation of WHO classification-based Prognostic Scoring System (WPSS) for myelodysplastic syndromes and comparison with the revised International Prognostic Scoring System (IPSS-R). A study of the International Working Group for Prognosis in Myelodyspla. Leukemia 2015, 29, 1502–1513. [Google Scholar] [CrossRef]
- Pfeilstöcker, M.; Tuechler, H.; Sanz, G.; Schanz, J.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Time-dependent changes in mortality and transformation risk in MDS. Blood 2016, 128, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Komrokji, R.S.; Corrales-Yepez, M.; Kharfan-Dabaja, M.A.; Al Ali, N.H.; Padron, E.; Rollison, D.E.; Pinilla-Ibarz, J.; Zhang, L.; Epling-Burnette, P.K.; Lancet, J.E.; et al. Hypoalbuminemia is an independent prognostic factor for overall survival in myelodysplastic syndromes. Am. J. Hematol. 2012, 87, 1006–1009. [Google Scholar] [CrossRef] [PubMed]
- van de Loosdrecht, A.A.; Ireland, R.; Kern, W.; Della Porta, M.G.; Alhan, C.; Balleisen, J.S.; Bettelheim, P.; Bowen, D.T.; Burbury, K.; Eidenschink, L.; et al. Rationale for the clinical application of flow cytometry in patients with myelodysplastic syndromes: Position paper of an International Consortium and the European LeukemiaNet Working Group. Leuk. Lymphoma 2013, 54, 472–475. [Google Scholar] [CrossRef]
- Chen-Liang, T.-H.; Casado-Prieto, A.M.; Campos-Rodríguez, V.; Hurtado, A.M.; Amigo, M.L.; García-Malo, M.D.; Vicente, V.; Ortuño, F.J.; Jerez, A. An increased percentage of myeloid CD34+ bone marrow cells stratifies intermediate IPSS-R myelodysplastic syndrome patients into prognostically significant groups. Int. J. Lab. Hematol. 2018, 40, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Calvo, X.; Arenillas, L.; Luño, E.; Senent, L.; Arnan, M.; Ramos, F.; Pedro, C.; Tormo, M.; Montoro, J.; Díez-Campelo, M.; et al. Enumerating bone marrow blasts from nonerythroid cellularity improves outcome prediction in myelodysplastic syndromes and permits a better definition of the intermediate risk category of the Revised International Prognostic Scoring System (IPSS-R). Am. J. Hematol. 2017, 92, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Porta, M.G.D.; Malcovati, L.; Boveri, E.; Travaglino, E.; Pietra, D.; Pascutto, C.; Passamonti, F.; Invernizzi, R.; Castello, A.; Magrini, U.; et al. Clinical relevance of bone marrow fibrosis and CD34-positive cell clusters in primary mvelodvsplastic syndromes. J. Clin. Oncol. 2009, 27, 754–762. [Google Scholar] [CrossRef]
- Fu, B.; Jaso, J.M.; Sargent, R.L.; Goswami, M.; Verstovsek, S.; Medeiros, L.J.; Wang, S.A. Bone marrow fibrosis in patients with primary myelodysplastic syndromes has prognostic value using current therapies and new risk stratification systems. Mod. Pathol. 2014, 27, 681–689. [Google Scholar] [CrossRef]
- Ramos, F.; Robledo, C.; Izquierdo-García, F.M.; Suárez-Vilela, D.; Benito, R.; Fuertes, M.; Insunza, A.; Barragán, E.; Del Rey, M.; García-Ruiz de Morales, J.M.; et al. Bone marrow fibrosis in myelodysplastic syndromes: A prospective evaluation including mutational analysis. Oncotarget 2016, 7, 30492–30503. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Makishima, H.; Yoshizato, T.; Yoshida, K.; Sekeres, M.A.; Radivoyevitch, T.; Suzuki, H.; Przychodzen, B.; Nagata, Y.; Meggendorfer, M.; Sanada, M.; et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017, 49, 204–212. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.S.; Garcia-Manero, G.; Barnard, J.; Roboz, G.J.; Steensma, D.P.; DeZern, A.E.; Zell, K.; Zimmerman, C.; Al Ali, N.; et al. The efficacy of current prognostic models in predicting outcome of patients with myelodysplastic syndromes at the time of hypomethylating agent failure. Haematologica 2016, 101, e224–e227. [Google Scholar] [CrossRef]
- Fenaux, P.; Platzbecker, U.; Ades, L. How we manage adults with myelodysplastic syndrome. Br. J. Haematol. 2020, 189, 1016–1027. [Google Scholar] [CrossRef]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical Effect of Point Mutations in Myelodysplastic Syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Sallman, D.A.; Komrokji, R.; Vaupel, C.; Cluzeau, T.; Geyer, S.M.; McGraw, K.L.; Al Ali, N.H.; Lancet, J.; McGinniss, M.J.; Nahas, S.; et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia 2016, 30, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, C.M.; Nazha, A.; Kneen, K.; Abazeed, M.E.; Meggendorfer, M.; Przychodzen, B.P.; Nadarajah, N.; Adema, V.; Nagata, Y.; Goyal, A.; et al. Consequences of mutant TET2 on clonality and subclonal hierarchy. Leukemia 2018, 32, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Roy, S.; Di Genua, C.; Burns, A.; McGraw, K.; Valletta, S.; Larrayoz, M.J.; Fernandez-Mercado, M.; Mason, J.; Killick, S.; et al. Targeted resequencing analysis of 31 genes commonly mutated in myeloid disorders in serial samples from myelodysplastic syndrome patients showing disease progression. Leukemia 2016, 30, 247–250. [Google Scholar] [CrossRef]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M.; et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014, 124, 1513–1521. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.I.; Jones, S.N. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl. Cancer Res. 2016, 5, 707–724. [Google Scholar] [CrossRef]
- Nazha, A.; Al-Issa, K.; Hamilton, B.K.; Radivoyevitch, T.; Gerds, A.T.; Mukherjee, S.; Adema, V.; Zarzour, A.; Abuhadra, N.; Patel, B.J.; et al. Adding molecular data to prognostic models can improve predictive power in treated patients with myelodysplastic syndromes. Leukemia 2017, 31, 2848–2850. [Google Scholar] [CrossRef]
- Montalban-Bravo, G.; Kanagal-Shamanna, R.; Benton, C.B.; Class, C.A.; Chien, K.S.; Sasaki, K.; Naqvi, K.; Alvarado, Y.; Kadia, T.M.; Ravandi, F.; et al. Genomic context and TP53 allele frequency define clinical outcomes in TP53-mutated myelodysplastic syndromes. Blood Adv. 2020, 4, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Haase, D.; Stevenson, K.E.; Neuberg, D.; Maciejewski, J.P.; Nazha, A.; Sekeres, M.A.; Ebert, B.L.; Garcia-Manero, G.; Haferlach, C.; Haferlach, T.; et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia 2019, 33, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Sebaa, A.; Ades, L.; Baran-Marzack, F.; Mozziconacci, M.-J.; Penther, D.; Dobbelstein, S.; Stamatoullas, A.; Récher, C.; Prebet, T.; Moulessehoul, S.; et al. Incidence of 17p deletions and TP53 mutation in myelodysplastic syndrome and acute myeloid leukemia with 5q deletion. Genes Chromosom. Cancer 2012, 51, 1086–1092. [Google Scholar] [CrossRef]
- Meggendorfer, M.; Haferlach, C.; Kern, W.; Haferlach, T. Molecular analysis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5 reveals a specific spectrum of molecular mutations with prognostic impact: A study on 123 patients and 27 genes. Haematologica 2017, 102, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Lodé, L.; Ménard, A.; Flet, L.; Richebourg, S.; Loirat, M.; Eveillard, M.; Le Bris, Y.; Godon, C.; Theisen, O.; Gagez, A.-L.; et al. Emergence and evolution of TP53 mutations are key features of disease progression in myelodysplastic patients with lower-risk del(5q) treated with lenalidomide. Haematologica 2018, 103, e143–e146. [Google Scholar] [CrossRef] [PubMed]
- Kulasekararaj, A.G.; Smith, A.E.; Mian, S.A.; Mohamedali, A.M.; Krishnamurthy, P.; Lea, N.C.; Gäken, J.; Pennaneach, C.; Ireland, R.; Czepulkowski, B.; et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br. J. Haematol. 2013, 160, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Vardiman, J.W.; Arber, D.A.; Orazi, A.; Hasserjian, R.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M. The 2016 revision to the World Health Organization classifi cation of myeloid neoplasms and acute leukemia. Blood 2016. [Google Scholar] [CrossRef]
- Malcovati, L.; Karimi, M.; Papaemmanuil, E.; Ambaglio, I.; Jädersten, M.; Jansson, M.; Elena, C.; Gallì, A.; Walldin, G.; Porta, M.G.D.; et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015, 126, 233–241. [Google Scholar] [CrossRef]
- Harada, H.; Harada, Y.; Niimi, H.; Kyo, T.; Kimura, A.; Inaba, T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood 2004, 103, 2316–2324. [Google Scholar] [CrossRef]
- Christiansen, D.H.; Andersen, M.K.; Pedersen-Bjergaard, J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood 2004, 104, 1474–1481. [Google Scholar] [CrossRef]
- Damm, F.; Chesnais, V.; Nagata, Y.; Yoshida, K.; Scourzic, L.; Okuno, Y.; Itzykson, R.; Sanada, M.; Shiraishi, Y.; Gelsi-Boyer, V.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 2013, 122, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Abuhadra, N.; Mukherjee, S.; Al-Issa, K.; Adema, V.; Hirsch, C.M.; Advani, A.; Przychodzen, B.; Makhoul, A.; Awada, H.; Maciejewski, J.P.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes (MDS): Clonal architecture and impact on outcomes. Leuk. Lymphoma 2019, 60, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Papaemmanuil, E.; Bowen, D.T.; Boultwood, J.; Della Porta, M.G.; Pascutto, C.; Travaglino, E.; Groves, M.J.; Godfrey, A.L.; Ambaglio, I.; et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011, 118, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1-mutant MDS as a distinct disease subtype: A proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-G.; Kim, H.-R.; Seo, B.-Y.; Lee, J.H.; Choi, S.-Y.; Kim, S.-H.; Shin, J.-H.; Suh, S.-P.; Ahn, J.-S.; Shin, M.-G. The prognostic impact of mutations in spliceosomal genes for myelodysplastic syndrome patients without ring sideroblasts. BMC Cancer 2015, 15, 484. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; 5Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 Mutation in Myelodysplasia with Ring Sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Thol, F.; Kade, S.; Schlarmann, C.; Löffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kölking, B.; Wichmann, M.; Görlich, K.; et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, O.; Gelsi-Boyer, V.; Cheok, M.; Grabar, S.; Della-Valle, V.; Picard, F.; Viguié, F.; Quesnel, B.; Beyne-Rauzy, O.; Solary, E.; et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood 2009, 114, 3285–3291. [Google Scholar] [CrossRef]
- Smith, A.E.; Mohamedali, A.M.; Kulasekararaj, A.; Lim, Z.Y.; Gäken, J.; Lea, N.C.; Przychodzen, B.; Mian, S.A.; Nasser, E.E.; Shooter, C.; et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 2010, 116, 3923–3932. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, S.-K.; Zou, Z.; Fan, R.-H.; Lyu, X.-D. Prognostic significance of TET2 mutations in myelodysplastic syndromes: A meta-analysis. Leuk. Res. 2017, 58, 102–107. [Google Scholar] [CrossRef]
- Santamaría, C.; Ramos, F.; Puig, N.; Barragán, E.; De Paz, R.; Pedro, C.; Insunza, A.; Tormo, M.; Del Cañizo, C.; Diez-Campelo, M.; et al. Simultaneous analysis of the expression of 14 genes with individual prognostic value in myelodysplastic syndrome patients at diagnosis: WT1 detection in peripheral blood adversely affects survival. Ann. Hematol. 2012, 91, 1887–1895. [Google Scholar] [CrossRef]
- Lin, T.-L.; Nagata, Y.; Kao, H.-W.; Sanada, M.; Okuno, Y.; Huang, C.-F.; Liang, D.-C.; Kuo, M.-C.; Lai, C.-L.; Lee, E.-H.; et al. Clonal leukemic evolution in myelodysplastic syndromes with TET2 and IDH1/2 mutations. Haematologica 2014, 99, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Damm, F.; Wichmann, M.; Göhring, G.; Schumann, C.; Bug, G.; Ottmann, O.; et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 2010, 95, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Hou, H.-A.; Chou, W.-C.; Kuo, Y.-Y.; Liu, C.-Y.; Chen, C.-Y.; Lai, Y.-J.; Tseng, M.-H.; Huang, C.-F.; Chiang, Y.-C.; et al. IDH mutations are closely associated with mutations of DNMT3A, ASXL1 and SRSF2 in patients with myelodysplastic syndromes and are stable during disease evolution. Am. J. Hematol. 2014, 89, 137–144. [Google Scholar] [CrossRef]
- Thol, F.; Friesen, I.; Damm, F.; Yun, H.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Chaturvedi, A.; Sharma, A.; Wichmann, M.; et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 2011, 29, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Thota, S.; Viny, A.D.; Makishima, H.; Spitzer, B.; Radivoyevitch, T.; Przychodzen, B.; Sekeres, M.A.; Levine, R.L.; Maciejewski, J.P. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 2014, 124, 1790–1798. [Google Scholar] [CrossRef] [PubMed]
- Paquette, R.; Landaw, E.; Pierre, R.; Kahan, J.; Lubbert, M.; Lazcano, O.; Isaac, G.; McCormick, F.; Koeffler, H. N-ras mutations are associated with poor prognosis and increased risk of leukemia in myelodysplastic syndrome. Blood 1993, 82, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.M.; Bejar, R.; Stevenson, K.; Neuberg, D.; Shi, Y.; Cubrich, C.; Richardson, K.; Eastlake, P.; Garcia-Manero, G.; Kantarjian, H.; et al. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia 2013, 27, 2077–2081. [Google Scholar] [CrossRef]
- Kao, H.W.; Sanada, M.; Liang, D.C.; Lai, C.L.; Lee, E.H.; Kuo, M.C.; Lin, T.L.; Shih, Y.S.; Wu, J.H.; Huang, C.F.; et al. A high occurrence of acquisition and/or expansion of C-CBL mutant clones in the progression of high-risk myelodysplastic syndrome to acute myeloid leukemia. Neoplasia 2011, 13, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Makishima, H.; Yoshida, K.; Nguyen, N.; Przychodzen, B.; Sanada, M.; Okuno, Y.; Ng, K.P.; Gudmundsson, K.O.; Vishwakarma, B.A.; Jerez, A.; et al. Somatic SETBP1 mutations in myeloid malignancies. Nat. Genet. 2013, 45, 942–946. [Google Scholar] [CrossRef]
- Inoue, D.; Kitaura, J.; Matsui, H.; Hou, H.-A.; Chou, W.-C.; Nagamachi, A.; Kawabata, K.C.; Togami, K.; Nagase, R.; Horikawa, S.; et al. SETBP1 mutations drive leukemic transformation in ASXL1-mutated MDS. Leukemia 2015, 29, 847–857. [Google Scholar] [CrossRef]
- Fernandez-Mercado, M.; Pellagatti, A.; Di Genua, C.; Larrayoz, M.J.; Winkelmann, N.; Aranaz, P.; Burns, A.; Schuh, A.; Calasanz, M.J.; Cross, N.C.P.; et al. Mutations in SETBP1 are recurrent in myelodysplastic syndromes and often coexist with cytogenetic markers associated with disease progression. Br. J. Haematol. 2013, 163, 235–239. [Google Scholar] [CrossRef]
- Damm, F.; Itzykson, R.; Kosmider, O.; Droin, N.; Renneville, A.; Chesnais, V.; Gelsi-Boyer, V.; De Botton, S.; Vey, N.; Preudhomme, C.; et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia 2013, 27, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, JCO2001659. [Google Scholar] [CrossRef]
- Menssen, A.J.; Walter, M.J. Genetics of progression from MDS to secondary leukemia. Blood 2020, 136, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Duncavage, E.J.; Jacoby, M.A.; Chang, G.S.; Miller, C.A.; Edwin, N.; Shao, J.; Elliott, K.; Robinson, J.; Abel, H.; Fulton, R.S.; et al. Mutation Clearance after Transplantation for Myelodysplastic Syndrome. N. Engl. J. Med. 2018, 379, 1028–1041. [Google Scholar] [CrossRef]
- Martín-Izquierdo, M.; Abáigar, M.; Hernández-Sánchez, J.M.; Tamborero, D.; López-Cadenas, F.; Ramos, F.; Lumbreras, E.; Madinaveitia-Ochoa, A.; Megido, M.; Labrador, J.; et al. Co-occurrence of cohesin complex and Ras signaling mutations during progression from myelodysplastic syndromes to secondary acute myeloid leukemia. Haematologica 2020. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.S.; Meggendorfer, M.; Mukherjee, S.; Al Ali, N.; Walter, W.; Hutter, S.; Padron, E.; Madanat, Y.F.; Sallman, D.A.; et al. A Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. Blood 2018, 132, 793. [Google Scholar] [CrossRef]
- Winter, S.; Shoaie, S.; Kordasti, S.; Platzbecker, U. Integrating the “Immunome” in the Stratification of Myelodysplastic Syndromes and Future Clinical Trial Design. J. Clin. Oncol. 2020, 38, 1723–1735. [Google Scholar] [CrossRef]
| Variable | Parameter | Score | Final Score | Risk Group | LFS Median (Years) | OS Median (Years) |
|---|---|---|---|---|---|---|
| Blasts in bone marrow (%) | <5 | 0 | 0 | Low | 9.4 | 5.7 |
| 5–10 | 0.5 | |||||
| 11–20 | 1.5 | |||||
| 21–30 | 2 | 0.5–1 | Intermediate-1 | 3.3 | 3.5 | |
| Cytogenetic aberrations | Normal, del(5q), del(20q) | 0 | ||||
| Other alterations | 0.5 | 1.5–2 | Intermediate-2 | 1.1 | 1.2 | |
| 3 or more alterations, Chrom 7 aberrations | 1 | |||||
| ≥2.5 | High | 0.2 | 0.4 | |||
| Number of cytopenias * | None or 1 | 0 | ||||
| 2 or 3 | 0.5 |
| Variable | Parameter | Score | Final Score | Risk Group | Cumulative Risk = 0.5 # | |
|---|---|---|---|---|---|---|
| OS (Month) | Time to AML (Month) | |||||
| WHO category | RA/RARS/5q– | 0 | 0 | Very low | 90 | NR |
| RCMD/RCMD-RS | 1 | |||||
| RAEB-1 | 2 | |||||
| 1 | Low | 66 | NR | |||
| RAEB-2 | 3 | |||||
| Cytogenetic aberrations | Normal, del(5q), del(20q) | 0 | ||||
| 2 | Intermediate | 42 | 32 | |||
| Other alterations | 1 | |||||
| 3 or more alterations, Chrom 7 aberrations | 2 | 3–4 | High | 30 | 24 | |
| 5–6 | Very high | 12 | 6 | |||
| Transfusion dependency * | No | 0 | ||||
| Regular | 1 | |||||
| Variable | Parameter | Score | Final Score | Risk Group | OS Median (Months) |
|---|---|---|---|---|---|
| Performance status | >2 | 2 | 0–4 | Low | 54 |
| Age, years | 60–64 | 1 | |||
| ≥65 | 2 | ||||
| Platelets, ×109/L | 50–199 | 1 | 5–6 | Intermediate 1 | 25 |
| 30–49 | 2 | ||||
| <30 | 3 | ||||
| Hemoglobin, g/dL | <12 | 2 | |||
| Bone marrow blasts, % | 5–10 | 1 | 7–8 | Intermediate 2 | 14 |
| 11–29 | 2 | ||||
| WBC, ×109/L | >20 | 2 | |||
| Karyotype | Chr 7 abnormalities or complex abnormalities (≥3) | 3 | 9–15 | High | 6 |
| Prior transfusion | Yes | 1 |
| Variable | Score | Final Score | Risk Group | Median Time to AML (Years) | OS, Median (Years) | ||
|---|---|---|---|---|---|---|---|
| Blasts in bone marrow (%) | <2 | 0 | ≤1.5 | Very low | NR | 8.8 | |
| >2 to <5 | 1 | ||||||
| 5–10 | 2 | ||||||
| >10 | 3 | ||||||
| Cytogenetic aberrations | −Y, del(11q) | 0 | |||||
| 2–3 | Low | 10.8 | 5.3 | ||||
| Normal, del(5q), del(12p), del(20q), double including del(5q) | 1 | ||||||
| del(7q), +8, +19, i(17q), any other single or double independent clones | 2 | ||||||
| 3.5–4.5 | Intermediate | 3.2 | 3 | ||||
| −7, inv(3)/t(3q)/del(3q), double including −7/del(7q), complex: 3 abnormalities | 3 | ||||||
| Complex: >3 abnormalities | 4 | 5–6 | High | 1.4 | 1.6 | ||
| Cytopenia | Hb (g/dL) | ≥10 | 0 | ||||
| 8–10 | 1 | ||||||
| <8 | 1.5 | ||||||
| Platelets (×109/L) | >100 | 0 | |||||
| ≥6.5 | Very High | 0.7 | 0.8 | ||||
| 50–<100 | 0.5 | ||||||
| <50 | 1 | ||||||
| ANC (×109/L) | >0.8 | 0 | |||||
| <0.8 | 0.5 | ||||||
| Pathway | Gene | Specific Group | Clinical Outcome | Reference | |||
|---|---|---|---|---|---|---|---|
| OS | Statistical Approach | Time to AML | Statistical Approach | ||||
| Transcription factors | TP53 | D | Mv | D | Mv | Nazhad et al. [29]; Haase et al. [31]; Bejar et al. [20] | |
| RUNX1 | D | Mv | C | - | Bejar et al. [20] | ||
| BCOR | All | N | Mv | N | Mv | Damm et al. [40]; Abuhadra et al. [41] | |
| Frameshift | D | Uv/Mv | D | Uv | |||
| RNA splicing | SF3B1 | Non-MDS-RS | C | Uv/Mv | C | Uv/Mv | Malcovati et al. [42,43]; Kang et al. [44] |
| MDS-RS | I | Mv | C | - | Papaemmanuil et al. [45] | ||
| SRSF2 | D | Mv | D | Mv | Thol et al. [46] | ||
| U2AF1 | D | Mv | C | - | Kang et al. [44] | ||
| DNA methylation | TET2 | All | C | Uv/Mv | C | Uv/Mv | Kosmider et.al. [47], Smith et al. [48], Guo et al. [49], Santamaría et al. [50] |
| High-risk | N | Mv | D | Mv | Lin et al. [51] | ||
| IDH1 | C | Uv | C | Uv | Thol et al. [52], Lin et al. [53] | ||
| IDH2 | D | Uv | C | - | Lin et al. [53] | ||
| Chromatin modifiers | EZH2 | D | Mv | N | Mv | Bejar et al. [20] | |
| ASXL1 | D | Mv | D | Mv | Bejar et al. [20], Thol et al. [54] | ||
| Cohesin complex | STAG2 | D | Mv | C | - | Thota et al. [55] | |
| RAS signaling | NRAS | C | Uv/Mv | D | Uv | Paquette et al. [56], Murphy et al. [57], Bejar et al. [20] | |
| CBL | N | Uv | C | - | Kao et al. [58] | ||
| Others | SETBP1 | D | Uv/Mv | D | Uv/Mv | Makishima et al. [59], Inoue et al. [60], Fernández-Mercado et al. [61], Damm et al. [62] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen-Liang, T.-H. Prognosis in Myelodysplastic Syndromes: The Clinical Challenge of Genomic Integration. J. Clin. Med. 2021, 10, 2052. https://doi.org/10.3390/jcm10102052
Chen-Liang T-H. Prognosis in Myelodysplastic Syndromes: The Clinical Challenge of Genomic Integration. Journal of Clinical Medicine. 2021; 10(10):2052. https://doi.org/10.3390/jcm10102052
Chicago/Turabian StyleChen-Liang, Tzu-Hua. 2021. "Prognosis in Myelodysplastic Syndromes: The Clinical Challenge of Genomic Integration" Journal of Clinical Medicine 10, no. 10: 2052. https://doi.org/10.3390/jcm10102052
APA StyleChen-Liang, T.-H. (2021). Prognosis in Myelodysplastic Syndromes: The Clinical Challenge of Genomic Integration. Journal of Clinical Medicine, 10(10), 2052. https://doi.org/10.3390/jcm10102052





