Proteomic Analyses of Vitreous in Proliferative Diabetic Retinopathy: Prior Studies and Future Outlook
Abstract
:1. Introduction
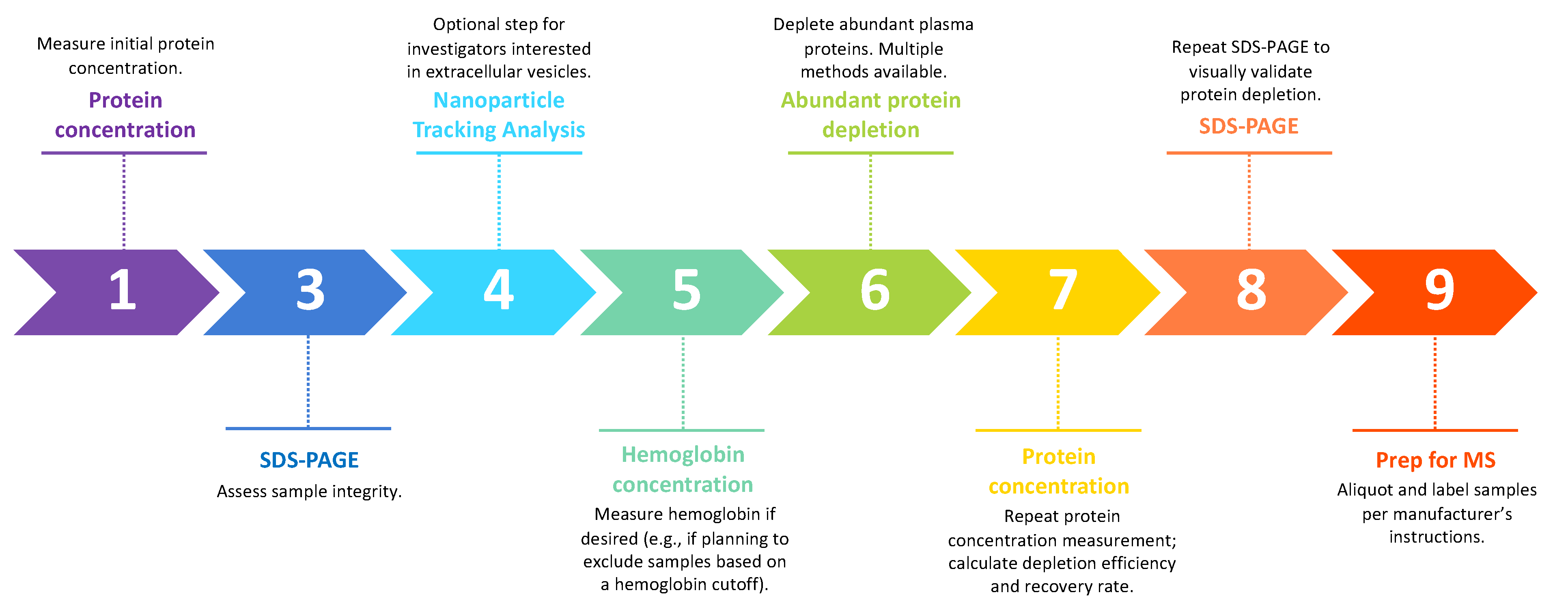
2. Workflow for Vitreous Proteomics
2.1. Sample Source and Selection
2.2. Storage
2.3. Sample Validation and Processing
2.3.1. Fractionation
2.3.2. Digestion
2.4. Mass Spectrometry
2.4.1. Mass Spectrometers: Basic Components
Ion Source
Mass Analyzers
2.5. Data Analysis
2.6. Differential Protein Expression Analysis
2.7. Data Publication
3. Prior Studies
3.1. Major Findings
3.2. Shortcomings
4. Moving the Field Forward
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sebag, J. Vitreous: In Health and Disease; Springer: New York, NY, USA, 2014. [Google Scholar]
- Bishop, P.N.; Crossman, M.V.; McLeod, D.; Ayad, S. Extraction and characterization of the tissue forms of collagen types II and IX from bovine vitreous. Biochem. J. 1994, 299, 497–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmut, O.; Mallinger, R.; Paschke, E. Studies on a distinct fraction of bovine vitreous body collagen. Graefes Arch. Klin. Exp. Ophthalmol. 1984, 221, 286–289. [Google Scholar] [CrossRef]
- Wu, C.W.; Sauter, J.L.; Johnson, P.K.; Chen, C.D.; Olsen, T.W. Identification and localization of major soluble vitreous proteins in human ocular tissue. Am. J. Ophthalmol. 2004, 137, 655–661. [Google Scholar]
- Zhao, Y.; Weber, S.R.; Lease, J.; Russo, M.; Siedlecki, C.A.; Xu, L.-C.; Chen, H.; Wang, W.; Ford, M.; Simó, R.; et al. Liquid Biopsy of Vitreous Reveals an Abundant Vesicle Population Consistent With the Size and Morphology of Exosomes. Transl. Vis. Sci. Technol. 2018, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angi, M.; Kalirai, H.; Coupland, S.; Damato, B.E.; Semeraro, F.; Romano, M.R. Proteomic Analyses of the Vitreous Humour. Mediat. Inflamm. 2012, 2012, 1–7, 148039. [Google Scholar] [CrossRef]
- Walia, S.; Clermont, A.C.; Gao, B.-B.; Aiello, L.P.; Feener, E.P. Vitreous proteomics and diabetic retinopathy. Semin. Ophthalmol. 2010, 25, 289–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, M.; Dacheva, I.; Nobl, M.; Siwy, J.; Schanstra, J.P.; Mullen, W.; Koch, F.H.J.; Kopitz, J.; Kretz, F.T.A.; Auffarth, G.U.; et al. Proteomic Analysis of Vitreous Humor in Retinal Vein Occlusion. PLoS ONE 2016, 11, e0158001. [Google Scholar] [CrossRef] [PubMed]
- Dacheva, I.; Reich, M.; Nobl, M.; Ceglowska, K.; Wasiak, J.; Siwy, J.; Zürbig, P.; Mischak, H.; Koch, F.H.J.; Kopitz, J.; et al. Proteome analysis of undiluted vitreous humor in patients with branch retinal vein occlusion. Ophthalmol. Z. Dtsch. Ophthalmol. Ges. 2018, 115, 203–215. [Google Scholar]
- Yu, J.; Peng, R.; Chen, H.; Cui, C.; Ba, J. Elucidation of the Pathogenic Mechanism of Rhegmatogenous Retinal Detachment with Proliferative Vitreoretinopathy by Proteomic Analysis. Investig. Ophthalmol. Vis. Sci. 2012, 53, 8146–8153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, L.M.; Santos, F.M.; Albuquerque, T.; Castro-De-Sousa, J.P.; Passarinha, L.A.; Tomaz, C.T. Proteome analysis of vitreous humor in retinal detachment using two different flow-charts for protein fractionation. J. Chromatogr. B 2017, 1061–1062, 334–341. [Google Scholar] [CrossRef]
- Naru, J.; Aggarwal, R.; Singh, U.; Mohanty, A.K.; Bansal, D.; Mangat, N.; Kakkar, N.; Agnihotri, N. Proteomic analysis of differentially expressed proteins in vitreous humor of patients with retinoblastoma using iTRAQ-coupled ESI-MS/MS approach. Tumour Biol. 2016, 37, 13915–13926. [Google Scholar] [CrossRef]
- Nobl, M.; Reich, M.; Dacheva, I.; Siwy, J.; Mullen, W.; Schanstra, J.P.; Choi, C.Y.; Kopitz, J.; Kretz, F.T.; Auffarth, G.; et al. Proteomics of vitreous in neovascular age-related macular degeneration. Exp. Eye Res. 2016, 146, 107–117. [Google Scholar] [CrossRef]
- Koss, M.J.; Hoffmann, J.; Nguyen, N.; Pfister, M.; Mischak, H.; Mullen, W.; Husi, H.; Rejdak, R.; Koch, F.; Jankowski, J.; et al. Proteomics of Vitreous Humor of Patients with Exudative Age-Related Macular Degeneration. PLoS ONE 2014, 9, e96895. [Google Scholar] [CrossRef]
- Sugioka, K.; Saito, A.; Kusaka, S.; Kuniyoshi, K.; Shimomura, Y. Identification of vitreous proteins in retinopathy of prematurity. Biochem. Biophys. Res. Commun. 2017, 488, 483–488. [Google Scholar] [CrossRef]
- Pfahler, S.M.; Brandford, A.N.; Glaser, B.M. A prospective study of in-office diagnostic vitreous sampling in patients with vitreoretinal pathology. Retina 2009, 29, 1032–1035. [Google Scholar] [CrossRef]
- Ghodasra, D.H.; Fante, R.; Gardner, T.W.; Langue, M.; Niziol, L.M.; Besirli, C.; Cohen, S.R.; Dedania, V.S.; Demirci, H.; Jain, N.; et al. Safety and Feasibility of Quantitative Multiplexed Cytokine Analysis From Office-Based Vitreous Aspiration. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3017–3023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skeie, J.M.; Roybal, C.N.; Mahajan, V.B. Proteomic Insight into the Molecular Function of the Vitreous. PLoS ONE 2015, 10, e0127567. [Google Scholar] [CrossRef]
- Gardner, T.W.; Sundstrom, J.M. A proposal for early and personalized treatment of diabetic retinopathy based on clinical pathophysiology and molecular phenotyping. Vis. Res. 2017, 139, 153–160. [Google Scholar] [CrossRef]
- Balazs, E.A. Fine Structure of the Developing Vitreous. Int. Ophthalmol. Clin. 1975, 15, 53–63. [Google Scholar] [CrossRef]
- Bishop, P.N. Structural macromolecules and supramolecular organisation of the vitreous gel. Prog. Retin. Eye Res. 2000, 19, 323–344. [Google Scholar] [CrossRef]
- Sebag, J. Ageing of the vitreous. Eye 1987, 1, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Sebag, J. Development and Aging of the Vitreous in the Vitreous: Structure, Function, and Pathobiology; Springer: New York, NY, USA, 1989; pp. 73–95. [Google Scholar]
- Neal, R.; Bettelheim, F.; Lin, C.; Winn, K.; Garland, D.; Zigler, J. Alterations in human vitreous humour following cataract extraction. Exp. Eye Res. 2005, 80, 337–347. [Google Scholar] [CrossRef]
- Mirzaei, M.; Gupta, V.B.; Chick, J.M.; Greco, T.M.; Wu, Y.; Chitranshi, N.; Wall, R.V.; Hone, E.; Deng, L.; Dheer, Y.; et al. Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci. Rep. 2017, 7, 1–16, 12685. [Google Scholar] [CrossRef] [Green Version]
- Schori, C.; Trachsel, C.; Grossmann, J.; Zygoula, I.; Barthelmes, D.; Grimm, C. The Proteomic Landscape in the Vitreous of Patients with Age-Related and Diabetic Retinal Disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD31–AMD40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasudhan, K.S.; Sarkar, S.; Gupta, V.; Gupta, A.; Chakraborti, A. Identification of unique proteins in vitreous fluid of patients with noninfectious uveitis. Acta Ophthalmol. 2018, 96, e989–e1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, C.; García-Ramírez, M.; Colomé, N.; Corraliza, L.; García-Pascual, L.; Casado, J.; Canals, F.; Simó, R. Identification of new pathogenic candidates for diabetic macular edema using fluorescence-based difference gel electrophoresis analysis. Diabetes Metab. Res. Rev. 2013, 29, 499–506. [Google Scholar] [CrossRef]
- Mostovenko, E.; Hassan, C.; Rattke, J.; Deelder, A.M.; Van Veelen, P.A.; Palmblad, M. Comparison of peptide and protein fractionation methods in proteomics. EuPA Open Proteom. 2013, 1, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Glatter, T.; Ludwig, C.; Ahrné, E.; Aebersold, R.; Heck, A.J.R.; Schmidt, A. Large-Scale Quantitative Assessment of Different In-Solution Protein Digestion Protocols Reveals Superior Cleavage Efficiency of Tandem Lys-C/Trypsin Proteolysis over Trypsin Digestion. J. Proteome Res. 2012, 11, 5145–5156. [Google Scholar] [CrossRef]
- Gross, J.H. Mass Spectrometry: A Textbook, 3rd ed.; Springer International Publishing AG: Cham, Switzerland, 2017. [Google Scholar]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- March, R.E. Ion Trap Mass Spectrometers in Encyclopedia of Spectroscopy and Spectrometry; Lindon, J.C., Tranter, G.E., Kopenaal, D.W., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; pp. 330–337. [Google Scholar]
- Schwartz, J.C.; Senko, M.W.; Syka, J.E.P. A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2002, 13, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Scigelova, M.; Hornshaw, M.; Giannakopulos, A.; Makarov, A. Fourier transform mass spectrometry. Mol. Cell. Proteom. MCP 2011, 10, M111.009431. [Google Scholar] [CrossRef] [Green Version]
- Marshall, A.G.; Hendrickson, C.L.; Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom. Rev. 1998, 17, 1–35. [Google Scholar] [CrossRef]
- Valaskovic, G.A.; Kelleher, N.L.; McLafferty, F.W. Attomole Protein Characterization by Capillary Electrophoresis-Mass Spectrometry. Science 1996, 273, 1199–1202. [Google Scholar] [CrossRef]
- Martin, S.E.; Shabanowitz, J.; Hunt, N.F.; Marto, J.A. Subfemtomole MS and MS/MS Peptide Sequence Analysis Using Nano-HPLC Micro-ESI Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2000, 72, 4266–4274. [Google Scholar] [CrossRef] [PubMed]
- Lipton, M.S.; Paša-Tolić, L.; Anderson, G.A.; Anderson, D.J.; Auberry, D.L.; Battista, J.R.; Daly, M.J.; Fredrickson, J.; Hixson, K.K.; Kostandarithes, H.; et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 2002, 99, 11049–11054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef] [Green Version]
- Bantscheff, M.; Schirle, M.; Sweetman, G.; Rick, J.; Kuster, B. Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. [Google Scholar] [CrossRef] [Green Version]
- Brenes, A.; Hukelmann, J.; Bensaddek, D.; Lamond, A.I. Multibatch TMT Reveals False Positives, Batch Effects and Missing Values. Mol. Cell. Proteom. 2019, 18, 1967–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutsch, E.W.; Mendoza, L.; Shteynberg, D.; Slagel, J.; Sun, Z.; Moritz, R.L. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteom. Clin. Appl. 2015, 9, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Eng, J.K.; Jahan, T.A.; Hoopmann, M.R. Comet: An open-source MS/MS sequence database search tool. Proteomics 2013, 13, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.T.; LePrevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LePrevost, F.D.V.; Haynes, S.E.; Avtonomov, D.M.; Chang, H.-Y.; Shanmugam, A.K.; Mellacheruvu, D.; Kong, A.T.; Nesvizhskii, A.I. Philosopher: A versatile toolkit for shotgun proteomics data analysis. Nat. Methods 2020, 17, 869–870. [Google Scholar] [CrossRef] [PubMed]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.; Forne, I.; Imhof, A. Bioinformatic analysis of proteomics data. BMC Syst. Biol. 2014, 8, S3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Chatr-Aryamontri, A.; Ceol, A.; Palazzi, L.M.; Nardelli, G.; Schneider, M.V.; Castagnoli, L.; Cesareni, G. MINT: The Molecular INTeraction database. Nucleic Acids Res. 2006, 35, D572–D574. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012, 40, D841–D846. [Google Scholar] [CrossRef]
- McDowall, M.; Scott, M.S.; Barton, G.J. PIPs: Human protein-protein interaction prediction database. Nucleic Acids Res. 2009, 37, D651–D656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.V.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Guo, N.; Kejariwal, A.; Thomas, P.D. PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007, 35, D247–D252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salomonis, N.; Hanspers, K.; Zambon, A.C.; Vranizan, K.; Lawlor, S.C.; Dahlquist, K.D.; Doniger, S.W.; Stuart, J.; Conklin, B.R.; Pico, A.R. GenMAPP 2: New features and resources for pathway analysis. BMC Bioinform. 2007, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larance, M.; Lamond, A. Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 2015, 16, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizcaíno, A.; Deutsch, J.; Wang, E.W.; Csordas, R.; Reisinger, A.; Ríos, F.D.; Dianes, A.J.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Alpi, E.; Wang, R.; Hermjakob, H.; Vizcaíno, J.A. Making proteomics data accessible and reusable: Current state of proteomics databases and repositories. Proteomics 2015, 15, 930–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, R.; Cortens, J.P.; Beavis, R.C. Open Source System for Analyzing, Validating, and Storing Protein Identification Data. J. Proteome Res. 2004, 3, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, A.K.; Yocum, A.K.; Nesvizhskii, A.I. Utility of RNA-seq and GPMDB Protein Observation Frequency for Improving the Sensitivity of Protein Identification by Tandem MS. J. Proteome Res. 2014, 13, 4113–4119. [Google Scholar] [CrossRef] [PubMed]
- Loukovaara, S.; Nurkkala, H.; Tamene, F.; Gucciardo, E.; Liu, X.; Repo, P.; Lehti, K.; Varjosalo, M. Quantitative Proteomics Analysis of Vitreous Humor from Diabetic Retinopathy Patients. J. Proteome Res. 2015, 14, 5131–5143. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Zhao, M.; Yu, J.; Zhu, D.; Wang, Y.; She, X.; Hu, Y.; Zheng, Z. Difference in the Vitreal Protein Profiles of Patients with Proliferative Diabetic Retinopathy with and without Intravitreal Conbercept Injection. J. Ophthalmol. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lu, Q.; Lu, P. Quantitative proteomics analysis of vitreous body from type 2 diabetic patients with proliferative diabetic retinopathy. BMC Ophthalmol. 2018, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, S.J.; Kim, K.; Kang, U.-B.; Lee, C.; Park, K.S.; Yu, H.G.; Kim, Y. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics 2007, 7, 4203–4215. [Google Scholar] [CrossRef]
- Gao, B.-B.; Chen, X.; Timothy, N.; Aiello, L.P.; Feener, E.P. Characterization of the Vitreous Proteome in Diabetes without Diabetic Retinopathy and Diabetes with Proliferative Diabetic Retinopathy. J. Proteome Res. 2008, 7, 2516–2525. [Google Scholar] [CrossRef]
- Gao, B.-B.; Clermont, A.; Rook, S.; Fonda, S.J.; Srinivasan, V.J.; Wojtkowski, M.; Fujimoto, J.G.; Avery, R.L.; Arrigg, P.G.; Bursell, S.-E.; et al. Extracellular carbonic anhydrase mediates hemorrhagic retinal and cerebral vascular permeability through prekallikrein activation. Nat. Med. 2007, 13, 181–188. [Google Scholar] [CrossRef]
- Koyama, R.; Nakanishi, T.; Ikeda, T.; Shimizu, A. Catalogue of soluble proteins in human vitreous humor by one-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrospray ionization mass spectrometry including seven angiogenesis-regulating factors. J. Chromatogr. B 2003, 792, 5–21. [Google Scholar] [CrossRef]
- Balaiya, S.; Zhou, Z.; Chalam, K.V. Characterization of Vitreous and Aqueous Proteome in Humans With Proliferative Diabetic Retinopathy and Its Clinical Correlation. Proteom. Insights 2017, 8, 1178641816686078. [Google Scholar] [CrossRef] [Green Version]
- Yamane, K.; Minamoto, A.; Yamashita, H.; Takamura, H.; Miyamoto-Myoken, Y.; Yoshizato, K.; Nabetani, T.; Tsugita, A.; Mishima, H.K. Proteome Analysis of Human Vitreous Proteins. Mol. Cell. Proteom. 2003, 2, 1177–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Feng, L.; Hu, J.W.; Xie, C.L.; Wang, F. Characterisation of the vitreous proteome in proliferative diabetic retinopathy. Proteome Sci. 2012, 10, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Kim, S.; Park, J.; Lee, H.K.; Park, K.S.; Yu, H.G.; Kim, Y. Differential expression of vitreous proteins in proliferative diabetic retinopathy. Curr. Eye Res. 2006, 31, 231–240. [Google Scholar] [CrossRef]
- Garcia-Ramírez, M.; Canals, F.; Hernández, C.; Colomé, N.; Ferrer, C.; Carrasco, E.; Garcia-Arumi, J.; Simó, R. Proteomic analysis of human vitreous fluid by fluorescence-based difference gel electrophoresis (DIGE): A new strategy for identifying potential candidates in the pathogenesis of proliferative diabetic retinopathy. Diabetologia 2007, 50, 1294–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavallée-Adam, M.; Rauniyar, N.; McClatchy, D.B.; Yates, J.R. PSEA-Quant: A Protein Set Enrichment Analysis on Label-Free and Label-Based Protein Quantification Data. J. Proteome Res. 2014, 13, 5496–5509. [Google Scholar] [CrossRef] [Green Version]
- Cha, S.; Imielinski, M.B.; Rejtar, T.; Richardson, E.A.; Thakur, D.; Sgroi, D.C.; Karger, B.L. In Situ Proteomic Analysis of Human Breast Cancer Epithelial Cells Using Laser Capture Microdissection: Annotation by Protein Set Enrichment Analysis and Gene Ontology. Mol. Cell. Proteom. 2010, 9, 2529–2544. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
| Publication | PMID | Experimental Groups | Total Number of Samples | Sample Size by Group | Mass Spectrometry | Instrument | Proteins Measured | Repository |
|---|---|---|---|---|---|---|---|---|
| Loukovaara, S., et al. Quantitative Proteomics Analysis of Vitreous Humor from Diabetic Retinopathy Patients. J. Proteome Res. 2015, 14 (12), 5131–5143. | 26490944 | PDR/NPDR/PDR (anti-VEGF)/NPDR (anti-VEGF) | 138 | 74 PDR, 49 NPDR, 5 PDR (anti-VEGF), 10 NPDR (anti-VEGF) | LC-MS/MS | Orbitrap Elite | 2482 | Peptide Atlas |
| Gardner, T.W., and Sundstrom, J.M. A proposal for early and personalized treatment of diabetic retinopathy based on clinical pathophysiology and molecular phenotyping. Vision Res. 2017. | 28438679 | PDR + NCVH/MH/ERM | 10 | 5 PDR + NCVH, 5 MH/ERM | Nano-LC-MS/MS | Q Exactive | 1213 | Supplemental |
| Zou, C., et al. Difference in the Vitreal Protein Profiles of Patients with Proliferative Diabetic Retinopathy with and without Intravitreal Conbercept Injection. J. Ophthalmol. 2018, 7397610. | 29850212 | PDR + IVC/PDR (no IVC)/MH | 26 | 9 PDR + IVC, 8 PDR (no IVC), 9 MH | LC-MS/MS | Not listed | 740 | Supplemental |
| Schori, C., et al. The Proteomic Landscape in the Vitreous of Patients with Age-Related and Diabetic Retinal Disease. Investig. Ophthalmol. Vis. Sci. 2018, 59 (4), AMD31–AMD40. | 30025106 | PDR/dry AMD/NV AMD/ERM | 34 | 9 PDR, 6 dry AMD, 10 NV AMD, 9 ERM | Nano-LC-MS/MS | Orbitrap Fusion | 677 | PRIDE |
| Li, J., et al. Quantitative proteomics analysis of vitreous body from type 2 diabetic patients with proliferative diabetic retinopathy. BMC Ophthalmol. 2018, 18 (1), 151. | 29940965 | PDR/MH | 18 | 9 PDR, 9 MH | LC-MS/MS | Orbitrap Elite | 610 | Full dataset not included |
| Kim, T., et al. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics 2007, 7 (22), 4203–4215. | 17955474 | PDR/MH | 33 | 11 PDR, 14 MH | IS/2-DE/MALDI-MS, nano-LC-MALDI-MS/MS, nano-LC-ESI-MS/MS | Thermo Electron model LTQ ESI linear single-quadrupole IT | 531 | Supplemental |
| Gao, B.-B., et al. Characterization of the Vitreous Proteome in Diabetes without Diabetic Retinopathy and Diabetes with Proliferative Diabetic Retinopathy. J. Proteome Res. 2008, 7, 2516–2525. | 18433156 | PDR/diabetic (no DR)/not diabetic | 17 | 7 PDR, 4 diabetic (no DR), 6 not diabetic | Nano-LC-MS/MS | LTQ Linear Ion Trap | 252 | Supplemental |
| Gao, B.-B., et al. Extracellular carbonic anhydrase mediates hemorrhagic retinal and cerebral vascular permeability through prekallikrein activation. Nat. Med. 2007, 13 (2), 181–188. | 17259996 | PDR/diabetic (no DR)/not diabetic | 25 | 13 PDR, 4 diabetic (no DR), 8 not diabetic | MS/MS | LTQ Linear Ion Trap | 117 | Supplemental |
| Koyama, R., et al. Catalogue of soluble proteins in human vitreous humor by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrospray ionization mass spectrometry including seven angiogenesis-regulating factors. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003, 792 (1), 5–21. | 12828993 | PDR | 1 | 1 PDR | ESI-IT-MS/MS | LCQDECA | 84 | Manuscript |
| Balaiya, S., et al. Characterization of Vitreous and Aqueous Proteome in Humans With Proliferative Diabetic Retinopathy and Its Clinical Correlation. Proteomics Insights 2017, 8, 1178641816686078. | 28469465 | PDR/MH/ERM | 10 | 5 PDR, 5 MH/ERM | Nano-LC-MS/MS | LTQ Orbitrap XL | 57 | Manuscript, supplemental |
| Yamane, K., et al. Proteome analysis of human vitreous proteins. Mol. Cell. Proteomics 2003, 2 (11), 1177–1187. | 12975481 | PDR/MH | 59 | 33 PDR, 26 MH | ESI-MS, MALDI-MS | Q-TOF, Voyager-DE STR | 38 | Manuscript |
| Wang, H., et al. Characterisation of the vitreous proteome in proliferative diabetic retinopathy. Proteome Sci. 2012, 10 (1), 15. | 22390717 | PDR/corneal transplant | 20 | 10 PDR, 10 corneal transplants | DIGE + MALDI-MS | Not listed | 29 | Full dataset not included |
| Hernandez, C., et al. Identification of new pathogenic candidates for diabetic macular edema using fluorescence-based difference gel electrophoresis analysis. Diabetes Metab. Res. Rev. 2013, 29 (6), 499–506. | 23568601 | PDR/DME/MH | 16 | 4 PDR, 4 DME, 8 MH | DIGE + MALDI-MS | Ultraflex | 25 | Full dataset not included |
| Kim, S.J., et al. Differential expression of vitreous proteins in proliferative diabetic retinopathy. Curr. Eye Res. 2006, 31 (3), 231–240. | 16531280 | PDR/MH | 30 | 15 PDR, 15 MH | 2-DE + MALDI-TOF, 2-DE + MS/MS | Not listed | 23 | Manuscript |
| Garcia-Ramirez, M., et al. Proteomic analysis of human vitreous fluid by fluorescence-based difference gel electrophoresis (DIGE): a new strategy for identifying potential candidates in the pathogenesis of proliferative diabetic retinopathy. Diabetologia 2007, 50 (6), 1294–1303. | 17380318 | PDR/MH | 18 | 8 PDR, 10 MH | DIGE + MALDI-MS | Ultraflex | 11 | Manuscript |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, S.R.; Zhao, Y.; Gates, C.; Ma, J.; da Veiga Leprevost, F.; Basrur, V.; Nesvizhskii, A.I.; Gardner, T.W.; Sundstrom, J.M. Proteomic Analyses of Vitreous in Proliferative Diabetic Retinopathy: Prior Studies and Future Outlook. J. Clin. Med. 2021, 10, 2309. https://doi.org/10.3390/jcm10112309
Weber SR, Zhao Y, Gates C, Ma J, da Veiga Leprevost F, Basrur V, Nesvizhskii AI, Gardner TW, Sundstrom JM. Proteomic Analyses of Vitreous in Proliferative Diabetic Retinopathy: Prior Studies and Future Outlook. Journal of Clinical Medicine. 2021; 10(11):2309. https://doi.org/10.3390/jcm10112309
Chicago/Turabian StyleWeber, Sarah R., Yuanjun Zhao, Christopher Gates, Jingqun Ma, Felipe da Veiga Leprevost, Venkatesha Basrur, Alexey I. Nesvizhskii, Thomas W. Gardner, and Jeffrey M. Sundstrom. 2021. "Proteomic Analyses of Vitreous in Proliferative Diabetic Retinopathy: Prior Studies and Future Outlook" Journal of Clinical Medicine 10, no. 11: 2309. https://doi.org/10.3390/jcm10112309







