NecroX-5 Can Suppress Melanoma Metastasis by Reducing the Expression of Rho-Family GTPases
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Cell Viability Assay
2.3. Cell Migration Assay
2.4. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.5. Immunohistochemistry
2.6. Western Blot
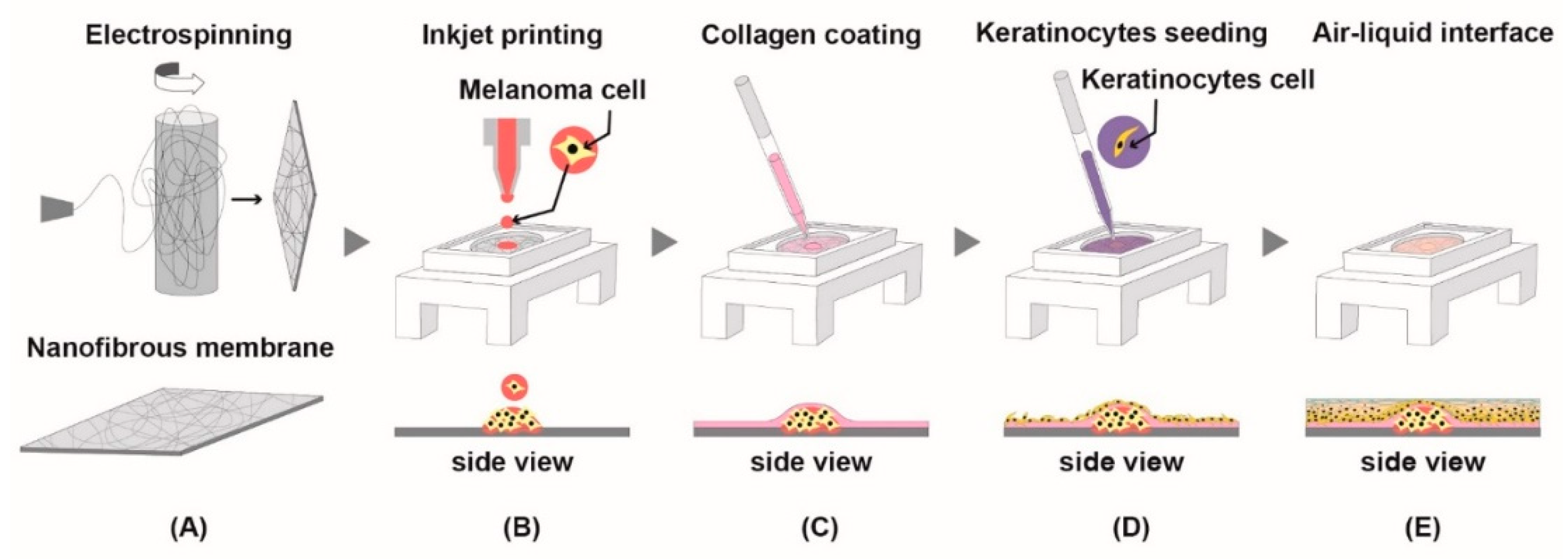
2.7. Fabrication of Nanofibrous Membrane
2.8. Printing Melanoma Cells and Aggregation
2.9. Development of Epidermis
2.10. Statistical Analysis
3. Results
3.1. Viability of Human Melanoma Cells after Treatment with NecroX-5
3.2. NecroX-5 Treatment Decreased Melanoma Cell Migration
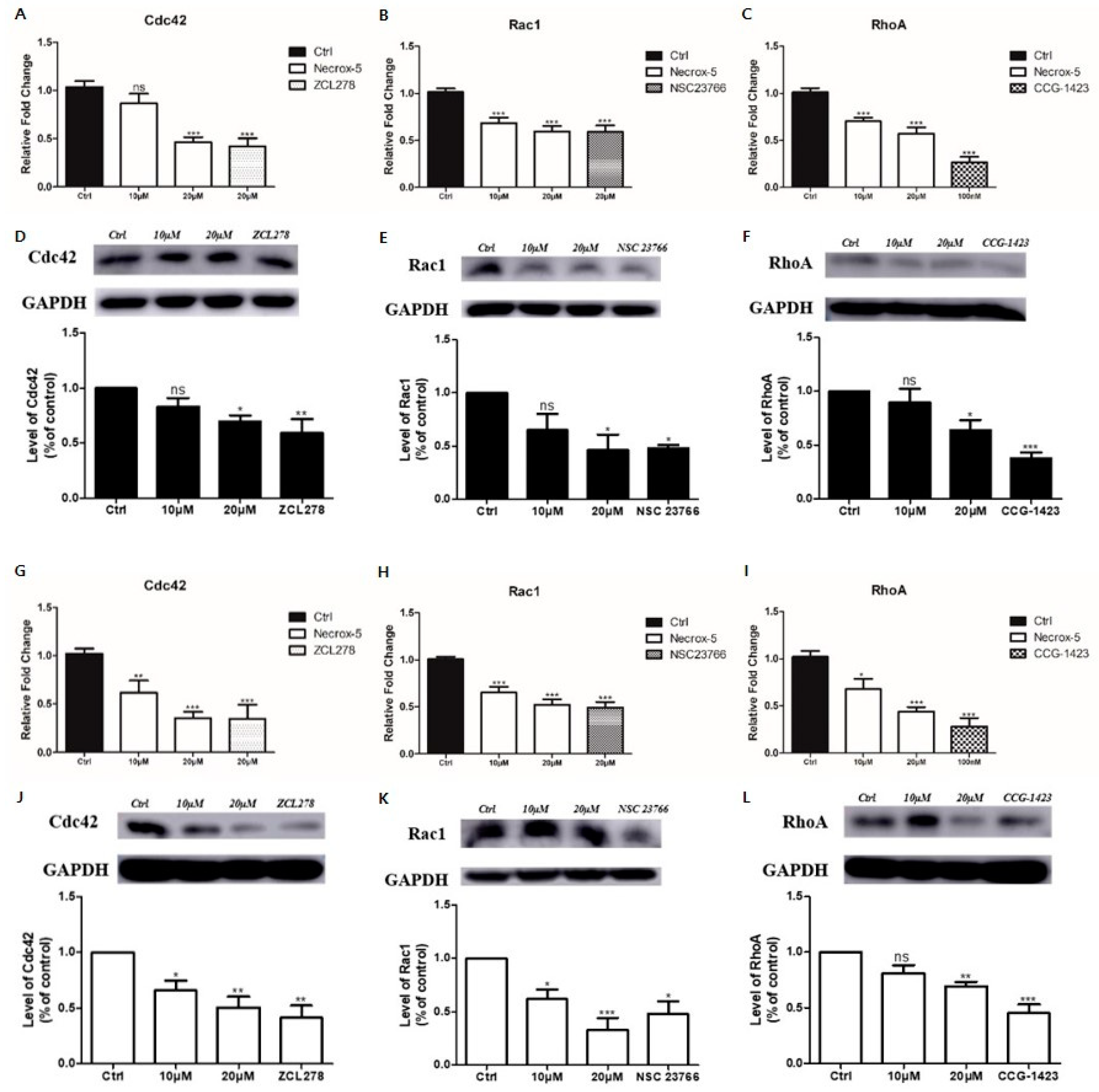
3.3. NecroX-5 Treatment Reduced Melanoma Cell Migration by Reducing the Expression of Rho-Family GTPases
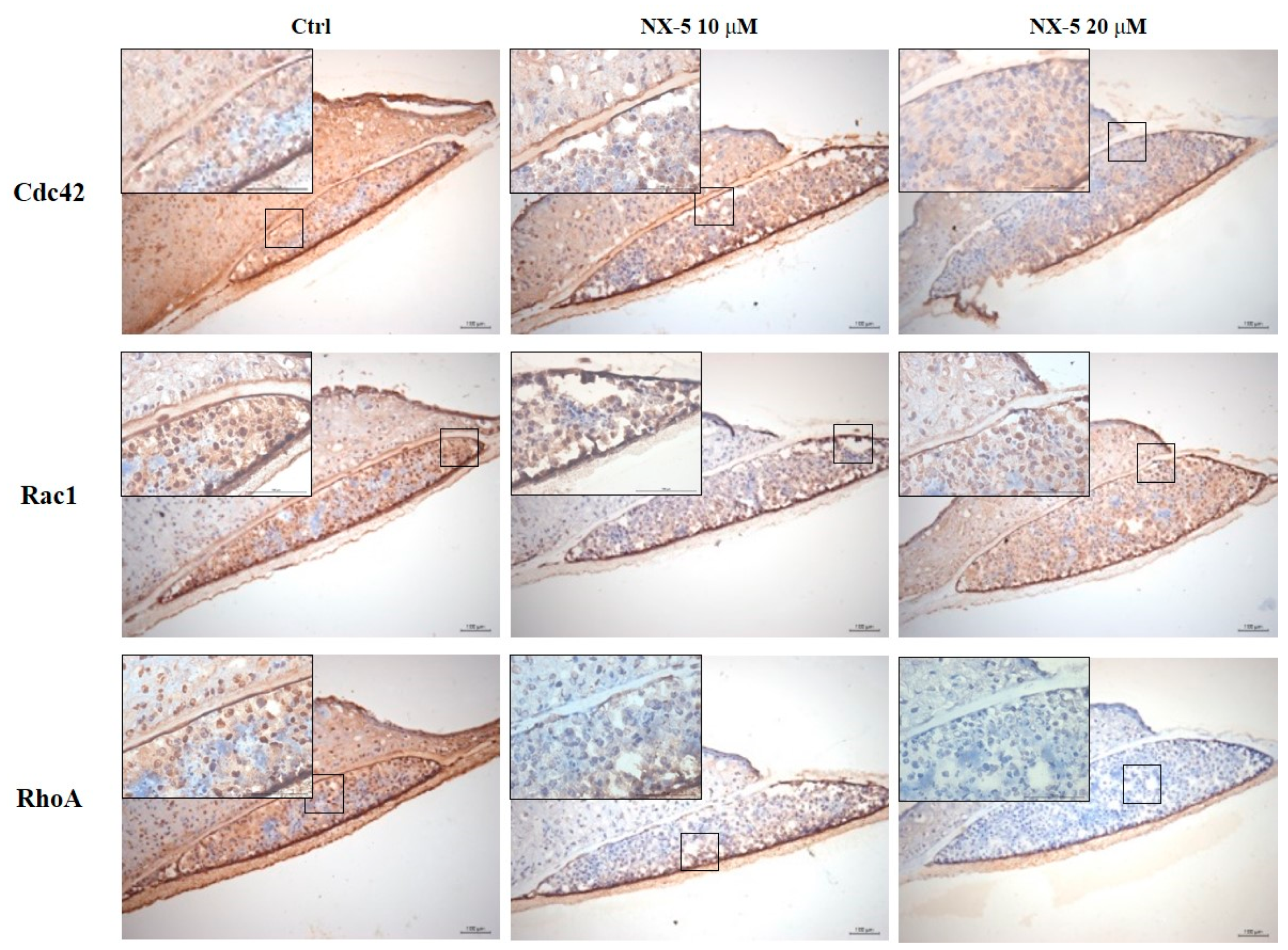
3.4. Necrox-5 Treatment Down-Regulated the Expression of Rho-Family GTPase in 3D Melanoma Model
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| EMT | Epithelial-mesenchymal transition |
| NX-5 | NecroX-5 |
References
- Scatena, C.; Murtas, D.; Tomei, S. Cutaneous melanoma classification: The importance of high-throughput genomic technologies. Front. Oncol. 2021, 11, 635488. [Google Scholar] [CrossRef]
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Tas, F. Metastatic behavior in melanoma: Timing, pattern, survival, and influencing factors. J. Oncol. 2012, 2012, 647684. [Google Scholar] [CrossRef]
- Domingues, B.; Lopes, J.M.; Soares, P.; Pópulo, H. Melanoma treatment in review. ImmunoTargets Ther. 2018, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef]
- Pedri, D.; Karras, P.; Landeloos, E.; Marine, J.C.; Rambow, F. Epithelial-to-mesenchymal-like transition events in melanoma. FEBS J. 2021. [Google Scholar] [CrossRef]
- Pearlman, R.L.; Montes de Oca, M.K.; Pal, H.C.; Afaq, F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017, 391, 125–140. [Google Scholar] [CrossRef]
- Vega, F.M.; Ridley, A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582, 2093–2101. [Google Scholar] [CrossRef]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef]
- Porter, A.P.; Papaioannou, A.; Malliri, A. Deregulation of Rho GTPases in cancer. Small GTPases 2016, 7, 123–138. [Google Scholar] [CrossRef]
- Gadea, G.; Sanz-Moreno, V.; Self, A.; Godi, A.; Marshall, C.J. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr. Biol. 2008, 18, 1456–1465. [Google Scholar] [CrossRef]
- Colón-Bolea, P.; García-Gómez, R.; Shackleton, S.; Crespo, P.; Bustelo, X.R.; Casar, B. RAC1 induces nuclear alterations through the LINC complex to enhance melanoma invasiveness. Mol. Biol. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Molina-Ortiz, I.; Bartolome, R.A.; Hernandez-Varas, P.; Colo, G.P.; Teixido, J. Overexpression of E-cadherin on melanoma cells inhibits chemokine-promoted invasion involving p190RhoGAP/p120ctn-dependent inactivation of RhoA. J. Biol. Chem. 2009, 284, 15147–15157. [Google Scholar] [CrossRef]
- Orgaz, J.L.; Sanz-Moreno, V. Emerging molecular targets in melanoma invasion and metastasis. Pigment Cell Melanoma Res. 2013, 26, 39–57. [Google Scholar] [CrossRef]
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef]
- Sahai, E.; Marshall, C.J. RHO-GTPases and cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef]
- Cho, S.G.; Keon-Il, I.; Lim, J.Y.; Nayoun, K.; Nam, Y.S.; Lee, E.S. Composition for Preventing or Treating Mucositis Comprising NecroX as Effective Ingredient. U.S. Patent 3,216,452, 14 August 2019. [Google Scholar]
- Park, J.H.; Kim, H.K.; Jung, H.; Kim, K.H.; Kang, M.S.; Hong, J.H.; Yu, B.C.; Park, S.; Seo, S.K.; Choi, I.W.; et al. NecroX-5 prevents breast cancer metastasis by AKT inhibition via reducing intracellular calcium levels. Int. J. Oncol. 2017, 50, 185–192. [Google Scholar] [CrossRef][Green Version]
- Park, T.M.; Kang, D.; Jang, I.; Yun, W.S.; Shim, J.H.; Jeong, Y.H.; Kwak, J.Y.; Yoon, S.; Jin, S. Fabrication of in vitro cancer microtissue array on fibroblast-layered nanofibrous membrane by inkjet printing. Int. J. Mol. Sci. 2017, 18, 2348. [Google Scholar] [CrossRef]
- Kang, D.; Kim, J.H.; Jeong, Y.H.; Kwak, J.Y.; Yoon, S.; Jin, S. Endothelial monolayers on collagen-coated nanofibrous membranes: Cell-cell and cell-ECM interactions. Biofabrication 2016, 8, 025008. [Google Scholar] [CrossRef]
- Jin, S.; Park, T.M.; Kim, C.H.; Kim, J.S.; Le, B.D.; Jeong, Y.H.; Kwak, J.Y.; Yoon, S. Three-dimensional migration of neutrophils through an electrospun nanofibrous membrane. Biotechniques 2015, 58, 285–292. [Google Scholar] [CrossRef]
- Kim, H.J.; Koo, S.Y.; Ahn, B.H.; Park, O.; Park, D.H.; Seo, D.O.; Won, J.H.; Yim, H.J.; Kwak, H.S.; Park, H.S.; et al. NecroX as a novel class of mitochondrial reactive oxygen species and ONOO(-) scavenger. Arch. Pharm. Res. 2010, 33, 1813–1823. [Google Scholar] [CrossRef]
- Fang, X.Z.; Ge, Y.L.; Chen, Z.Y.; Shu, H.Q.; Yang, Y.Y.; Yu, Y.; Zhou, X.J.; Chen, L.; Cui, S.N.; Wang, Y.X.; et al. NecroX-5 alleviate lipopolysaccharide-induced acute respiratory distress syndrome by inhibiting TXNIP/NLRP3 and NF-kappaB. Int. Immunopharmacol. 2020, 81, 106257. [Google Scholar] [CrossRef]
- Nam, Y.S.; Im, K.I.; Kim, N.; Song, Y.; Lee, J.S.; Jeon, Y.W.; Cho, S.G. Down-regulation of intracellular reactive oxygen species attenuates P-glycoprotein-associated chemoresistance in Epstein-Barr virus-positive NK/T-cell lymphoma. Am. J. Transl. Res. 2019, 11, 1359–1373. [Google Scholar]
- Ju, R.J.; Stehbens, S.J.; Haass, N.K. The role of melanoma cell-stroma interaction in cell motility, invasion, and metastasis. Front. Med. 2018, 5, 307. [Google Scholar] [CrossRef]
- Parri, M.; Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun. Sign. 2010, 8, 23. [Google Scholar] [CrossRef]
- Ungefroren, H.; Witte, D.; Lehnert, H. The role of small GTPases of the Rho/Rac family in TGF-beta-induced EMT and cell motility in cancer. Dev. Dyn. 2018, 247, 451–461. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Lionarons, D.A.; Hancock, D.C.; Rana, S.; East, P.; Moore, C.; Murillo, M.M.; Carvalho, J.; Spencer-Dene, B.; Herbert, E.; Stamp, G.; et al. RAC1(P29S) induces a mesenchymal phenotypic switch via serum response factor to promote melanoma development and therapy resistance. Cancer Cell 2019, 36, 68–83.e69. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.D.M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed]
- Dua, P.; Gude, R.P. Pentoxifylline impedes migration in B16F10 melanoma by modulating Rho GTPase activity and actin organisation. Eur. J. Cancer 2008, 44, 1587–1595. [Google Scholar] [CrossRef]
- Friesland, A.; Zhao, Y.; Chen, Y.H.; Wang, L.; Zhou, H.; Lu, Q. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. USA 2013, 110, 1261–1266. [Google Scholar] [CrossRef]
- Maldonado, M.D.M.; Medina, J.I.; Velazquez, L.; Dharmawardhane, S. Targeting Rac and Cdc42 GEFs in metastatic cancer. Front. Cell Dev. Biol. 2020, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Evelyn, C.R.; Bell, J.L.; Ryu, J.G.; Wade, S.M.; Kocab, A.; Harzdorf, N.L.; Showalter, H.D.; Neubig, R.R.; Larsen, S.D. Design, synthesis and prostate cancer cell-based studies of analogs of the Rho/MKL1 transcriptional pathway inhibitor, CCG-1423. Bioorg. Med. Chem. Lett. 2010, 20, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Bourland, J.; Fradette, J.; Auger, F.A. Tissue-engineered 3D melanoma model with blood and lymphatic capillaries for drug development. Sci. Rep. 2018, 8, 13191. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, G.-T.; Lee, J.-H.; Jeong, S.-H.; Jin, S.-W.; Park, Y.-M. NecroX-5 Can Suppress Melanoma Metastasis by Reducing the Expression of Rho-Family GTPases. J. Clin. Med. 2021, 10, 2790. https://doi.org/10.3390/jcm10132790
Moon G-T, Lee J-H, Jeong S-H, Jin S-W, Park Y-M. NecroX-5 Can Suppress Melanoma Metastasis by Reducing the Expression of Rho-Family GTPases. Journal of Clinical Medicine. 2021; 10(13):2790. https://doi.org/10.3390/jcm10132790
Chicago/Turabian StyleMoon, Gue-Tae, Ji-Hyun Lee, Sang-Hyun Jeong, Song-Wan Jin, and Young-Min Park. 2021. "NecroX-5 Can Suppress Melanoma Metastasis by Reducing the Expression of Rho-Family GTPases" Journal of Clinical Medicine 10, no. 13: 2790. https://doi.org/10.3390/jcm10132790
APA StyleMoon, G.-T., Lee, J.-H., Jeong, S.-H., Jin, S.-W., & Park, Y.-M. (2021). NecroX-5 Can Suppress Melanoma Metastasis by Reducing the Expression of Rho-Family GTPases. Journal of Clinical Medicine, 10(13), 2790. https://doi.org/10.3390/jcm10132790





