+3179G/A Insulin-Like Growth Factor-1 Receptor Polymorphism: A Novel Susceptibility Contributor in Anti-Ro/SSA Positive Patients with Sjögren’s Syndrome: Potential Clinical and Pathogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA Extraction
2.3. Restriction Fragment Length Polymorphism–Polymerase Chain Reaction (RFLP–PCR)
2.4. cDNA Synthesis and Real-Time PCR
2.5. Immunohistochemistry
2.6. Statistics
3. Results
3.1. Clinical and Serological Characteristics of Study Participants
3.2. Allele and Genotype Analysis in SS, SS-Lymphoma Patients, and HCs
3.3. Clinical and Laboratory Associations of the IGF1R Polymorphism rs2229765 among SS Patients
3.4. Allele and Genotype Analyses in SS, SS-Lymphoma Patients According to Anti-Ro/SSA Status and HCs
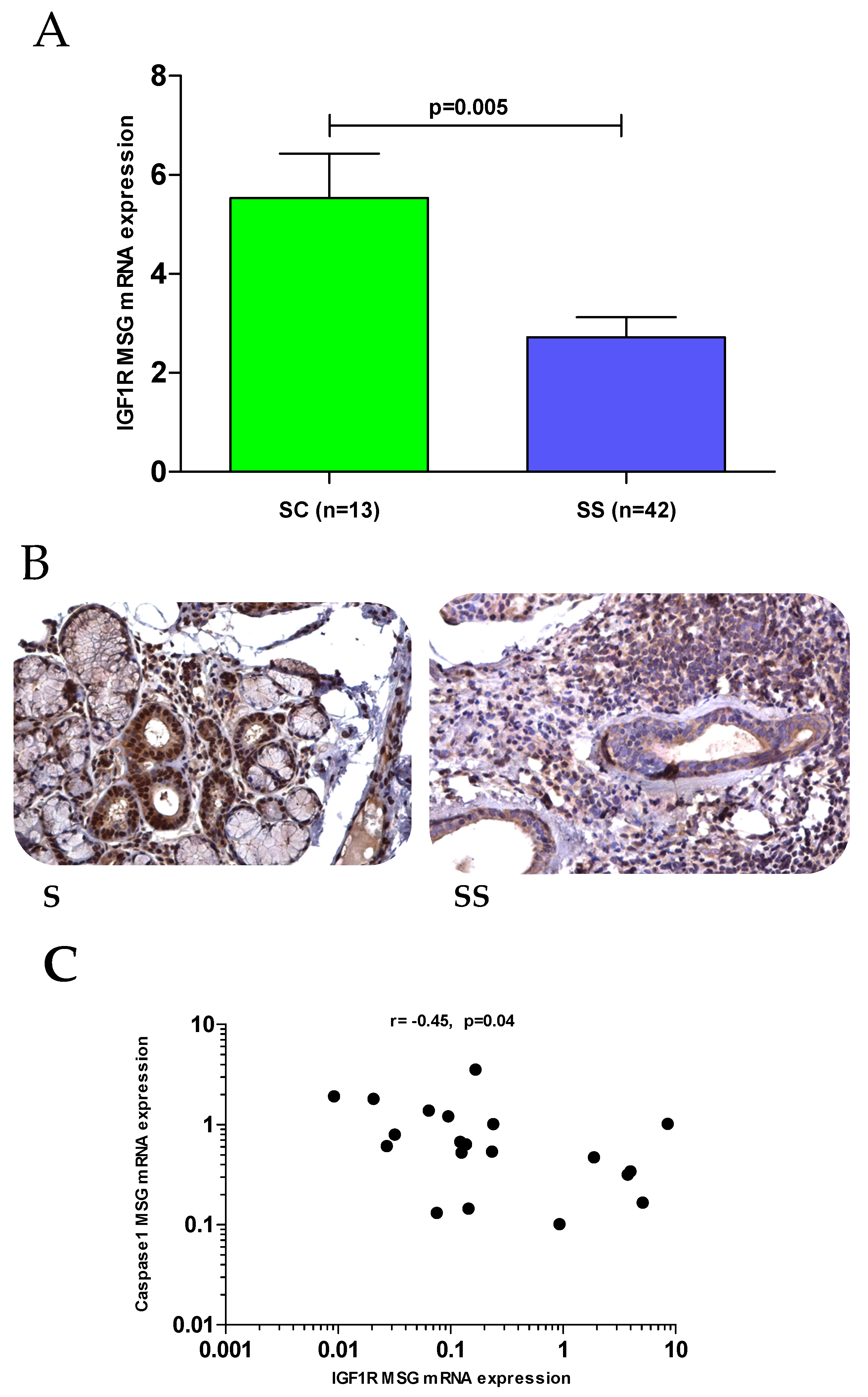
3.5. Gene and Protein Expression of IGF1/IGF1R Axis Components in MSG Tissues
3.6. Dampened IGF1R Expression and Apoptotic Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mavragani, C.P.; Moutsopoulos, H.M. Sjögren Syndrome Review. Can. Med. Assoc. J. 2014, 186, 579–586. [Google Scholar] [CrossRef]
- Mariette, X.; Criswell, L.A. Primary Sjögren’s Syndrome. N. Engl. J. Med. 2018, 378, 931–939. [Google Scholar] [CrossRef]
- Qin, B.; Wang, J.; Yang, Z.; Yang, M.; Ma, N.; Huang, F.; Zhong, R. Epidemiology of primary Sjögren’s syndrome: A systematic review and meta-analysis. Ann. Rheum. Dis. 2015, 74, 1983–1989. [Google Scholar] [CrossRef]
- Nezos, A.; Gravani, F.; Tassidou, A.; Kapsogeorgou, E.K.; Voulgarelis, M.; Koutsilieris, M.; Crow, M.K.; Mavragani, C.P. Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: Contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis. J. Autoimmun. 2015, 63, 47–58. [Google Scholar] [CrossRef]
- Bodewes, I.L.A.; Versnel, M.A. Interferon activation in primary Sjögren’s syndrome: Recent insights and future perspective as novel treatment target. Expert Rev. Clin. Immunol. 2018, 14, 817–829. [Google Scholar] [CrossRef]
- Huijser, E.; Versnel, M. Making Sense of Intracellular Nucleic Acid Sensing in Type I Interferon Activation in Sjögren’s Syndrome. J. Clin. Med. 2021, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Cinoku, I.I.; Verrou, K.-M.; Piperi, E.; Voulgarelis, M.; Moutsopoulos, H.M.; Mavragani, C.P. Interferon (IFN)-stimulated gene 15: A novel biomarker for lymphoma development in Sjögren’s syndrome. J. Autoimmun. 2021, 123, 102704. [Google Scholar] [CrossRef] [PubMed]
- Zintzaras, E.; Voulgarelis, M.; Moutsopoulos, H.M. The Risk of Lymphoma Development in Autoimmune Diseases: A Meta-Analysis. Arch. Intern. Med. 2005, 165, 2337–2344. [Google Scholar] [CrossRef]
- Stergiou, I.E.; Poulaki, A.; Voulgarelis, M. Pathogenetic Mechanisms Implicated in Sjögren’s Syndrome Lymphomagenesis: A Review of the Literature. J. Clin. Med. 2020, 9, 3794. [Google Scholar] [CrossRef]
- Skarlis, C.; Argyriou, E.; Mavragani, C.P. Lymphoma in Sjögren’s Syndrome: Predictors and Therapeutic Options. Curr. Treat. Options Rheumatol. 2020, 6, 1–17. [Google Scholar] [CrossRef]
- Verstappen, G.M.; Pringle, S.; Bootsma, H.; Kroese, F.G.M. Epithelial–immune cell interplay in primary Sjögren syndrome salivary gland pathogenesis. Nat. Rev. Rheumatol. 2021, 17, 333–348. [Google Scholar] [CrossRef]
- Psianou, K.; Panagoulias, I.; Papanastasiou, A.D.; De Lastic, A.-L.; Rodi, M.; Spantidea, P.I.; Degn, S.E.; Georgiou, P.; Mouzaki, A. Clinical and immunological parameters of Sjögren’s syndrome. Autoimmun. Rev. 2018, 17, 1053–1064. [Google Scholar] [CrossRef]
- Mavragani, C.P.; Fragoulis, G.E.; Moutsopoulos, H.M. Endocrine alterations in primary Sjogren’s syndrome: An overview. J. Autoimmun. 2012, 39, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Sisto, M.; Lisi, S.; Lofrumento, D.D.; D’Amore, M.; Scagliusi, P.; Mitolo, V. Autoantibodies from Sjogren’s Syndrome Trigger Apoptosis in Salivary Gland Cell Line. Ann. N. Y. Acad. Sci. 2007, 1108, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, P.; Fietta, P. Apoptosis and Sjögren syndrome. Semin. Arthritis Rheum. 2003, 33, 49–65. [Google Scholar] [CrossRef]
- Polihronis, M.; Tapinos, N.I.; Theocharis, S.E.; Economou, A.; Kittas, C.; Moutsopoulos, H.M. Modes of epithelial cell death and repair in Sjögren’s syndrome (SS). Clin. Exp. Immunol. 1998, 114, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Manoussakis, M.N.; Boiu, S.; Korkolopoulou, P.; Kapsogeorgou, E.K.; Kavantzas, N.; Ziakas, P.; Patsouris, E.; Moutsopoulos, H.M. Rates of infiltration by macrophages and dendritic cells and expression of interleukin-18 and interleukin-12 in the chronic inflammatory lesions of Sjögren’s syndrome: Correlation with certain features of immune hyperactivity and factors associated with high risk of lymphoma development. Arthritis Rheum. 2007, 56, 3977–3988. [Google Scholar] [CrossRef]
- Vakrakou, A.G.; Boiu, S.; Ziakas, P.; Xingi, E.; Boleti, H.; Manoussakis, M.N. Systemic activation of NLRP3 inflammasome in patients with severe primary Sjögren’s syndrome fueled by inflammagenic DNA accumulations. J. Autoimmun. 2018, 91, 23–33. [Google Scholar] [CrossRef]
- Baldini, C.; Santini, E.; Rossi, C.; Donati, V.; Solini, A. The P2X7 receptor-NLRP3 inflammasome complex predicts the development of non-Hodgkin’s lymphoma in Sjogren’s syndrome: A prospective, observational, single-centre study. J. Intern. Med. 2017, 282, 175–186. [Google Scholar] [CrossRef]
- Blokland, S.L.M.; Flessa, C.-M.; van Roon, J.A.G.; Mavragani, C.P. Emerging roles for chemokines and cytokines as orchestrators of immunopathology in Sjögren’s syndrome. Rheumatology 2019, 60, 3072–3087. [Google Scholar] [CrossRef]
- Adams, T.E.; Epa, V.C.; Garrett, T.P.J.; Ward, C.W. Structure and function of the type 1 insulin-like growth factor receptor. Cell. Mol. Life Sci. 2000, 57, 1050–1093. [Google Scholar] [CrossRef]
- Hjortebjerg, R.; Frystyk, J. Determination of IGFs and their binding proteins. Best Pract. Res. Clin. Endocrinol. Metab. 2013, 27, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Hakuno, F.; Takahashi, S.-I. 40 YEARS OF IGF1: IGF1 receptor signaling pathways. J. Mol. Endocrinol. 2018, 61, T69–T86. [Google Scholar] [CrossRef]
- Mitsui, R.; Fujita-Yoshigaki, J.; Narita, T.; Matsuki-Fukushima, M.; Satoh, K.; Qi, B.; Guo, M.-Y.; Katsumata-Kato, O.; Sugiya, H. Maintenance of paracellular barrier function by insulin-like growth factor-I in submandibular gland cells. Arch. Oral Biol. 2010, 55, 963–969. [Google Scholar] [CrossRef]
- Skarlis, C.; Nezos, A.; Mavragani, C.P.; Koutsilieris, M. The role of insulin growth factors in autoimmune diseases. Ann. Res. Hosp. 2019, 3. [Google Scholar] [CrossRef]
- Mustafa, W.; Mustafa, A.; Elbakri, N.; Link, H.; Adem, A. Reduced Levels of Insulin-Like Growth Factor-1 Receptor (IGF-1R) Suppress Cellular Signaling in Experimental Autoimmune Sialadenitis (EAS). J. Recept. Signal Transduct. Res. 2001, 21, 47–54. [Google Scholar] [CrossRef]
- Katz, J.; Stavropoulos, F.; Cohen, D.; Robledo, J.; Stewart, C.; Heft, M. IGF-1 and insulin receptor expression in the minor salivary gland tissues of Sjögren’s syndrome and mucoceles—Immunohistochemical study. Oral Dis. 2003, 9, 7–13. [Google Scholar] [CrossRef]
- Markopoulos, A.K.; Poulopoulos, A.K.; Kayavis, I.; Papanayotou, P. Immunohistochemical detection of insulin-like growth factor-I in the labial salivary glands of patients with Sjögren’s syndrome. Oral Dis. 2000, 6, 31–34. [Google Scholar] [CrossRef]
- Emamian, E.S.; Leon, J.M.; Lessard, C.J.; Grandits, M.; Baechler, E.C.; Gaffney, P.; Segal, B.; Rhodus, N.L.; Moser, K.L. Peripheral blood gene expression profiling in Sjögren’s syndrome. Genes Immun. 2009, 10, 285–296. [Google Scholar] [CrossRef]
- Stanilova, S.; Ivanova, M.; Karakolev, I.; Stoilov, R.; Rashkov, R.; Manolova, I.; Stanilova, S. Association of +3179G/A insulin-like growth factor-1 receptor polymorphism and insulin-like growth factor-1 serum level with systemic lupus erythematosus. Lupus 2013, 22, 1388–1393. [Google Scholar] [CrossRef]
- Stanilov, N.S.; Karakolev, I.A.; Deliysky, T.S.; Jovchev, J.P.; Stanilova, S. Association of insulin-like growth factor-I receptor polymorphism with colorectal cancer development. Mol. Biol. Rep. 2014, 41, 8099–8106. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Shiboski, S.C.; Seror, R.; A Criswell, L.; Labetoulle, M.; Lietman, T.M.; Rasmussen, A.; Scofield, H.; Vitali, C.; Bowman, S.J.; et al. 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjögren’s syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Ann. Rheum. Dis. 2016, 76, 9–16. [Google Scholar] [CrossRef]
- Quintanilla-Martinez, L. The 2016 updated WHO classification of lymphoid neoplasias. Hematol. Oncol. 2017, 35, 37–45. [Google Scholar] [CrossRef]
- Argyriou, E.; Nezos, A.; Roussos, P.; Venetsanopoulou, A.; Voulgarelis, M.; Boki, K.; Tzioufas, A.; Moutsopoulos, H.; Mavragani, C. Leukocyte Immunoglobulin-Like Receptor A3 (LILRA3): A Novel Marker for Lymphoma Development among Patients with Young Onset Sjogren’s Syndrome. J. Clin. Med. 2021, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Meyerholz, D.K.; Beck, A.P. Principles and approaches for reproducible scoring of tissue stains in research. Lab. Investig. 2018, 98, 844–855. [Google Scholar] [CrossRef]
- Papageorgiou, A.; Mavragani, C.P.C.P.; Nezos, A.; Zintzaras, E.; Quartuccio, L.; De Vita, S.; Koutsilieris, M.; Tzioufas, A.G.A.G.; Moutsopoulos, H.M.; Voulgarelis, M. A BAFF Receptor His159Tyr Mutation in Sjögren’s Syndrome-Related Lymphoproliferation. Arthritis Rheumatol. 2015, 67, 2732–2741. [Google Scholar] [CrossRef] [PubMed]
- Denley, A.; Cosgrove, L.J.; Booker, G.W.; Wallace, J.C.; Forbes, B.E. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005, 16, 421–439. [Google Scholar] [CrossRef]
- Kooijman, R. Regulation of apoptosis by insulin-like growth factor (IGF)-I. Cytokine Growth Factor Rev. 2006, 17, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulou, F.; Petraki, C.; Philippou, A.; Analitis, A.; Msaouel, P.; Koutsilieris, M. Expression of IGF-IEc Isoform in Renal Cell Carcinoma Tissues. Anticancer. Res. 2020, 40, 6213–6219. [Google Scholar] [CrossRef] [PubMed]
- Mitsias, D.I.; Kapsogeorgou, E.K.; Moutsopoulos, H.M. The role of epithelial cells in the initiation and perpetuation of autoimmune lesions: Lessons from Sjögren’s syndrome (autoimmune epithelitis). Lupus 2006, 15, 255–261. [Google Scholar] [CrossRef]
- Benchabane, S.; Slimani-Kaddouri, A.; Acheli, D.; Bendimerad-Iratene, T.; Mesbah, R.; Touil-Boukoffa, C. Association between increased Bcl-2, Fas and FasL levels and inflammation extent in labial salivary glands during primary Sjögren’s syndrome. Endocr. Metab. Immune Disord. Drug Targets 2021. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Warner, B.M.; Odani, T.; Ji, Y.; Mo, Y.-Q.; Nakamura, H.; Jang, S.-I.; Yin, H.; Michael, D.G.; Hirata, N.; et al. LAMP3 induces apoptosis and autoantigen release in Sjögren’s syndrome patients. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Kapsogeorgou, E.K.; Abu-Helu, R.F.; Moutsopoulos, H.M.; Manoussakis, M.N. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005, 52, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Choe, J.-Y.; Lee, G.H. Enhanced expression of NLRP3 inflammasome-related inflammation in peripheral blood mononuclear cells in Sjögren’s syndrome. Clin. Chim. Acta 2017, 474, 147–154. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Manipulation of Host Cell Death Pathways during Microbial Infections. Cell Host Microbe 2010, 8, 44–54. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.; Weiss, H.; Goldman, B.; Kanety, H.; Stannard, B.; Leroith, D.; Shemer, J. Cytokines and growth factors modulate cell growth and insulin-like growth factor binding protein secretion by the human salivary cell line (HSG). J. Cell. Physiol. 1995, 165, 223–227. [Google Scholar] [CrossRef]
- Katz, J.; Nasatzky, E.; Werner, H.; Roith, D.; Shemer, J. Tumor necrosis factor α and interferon γ–induced cell growth arrest is mediated via insulin-like growth factor binding protein-3. Growth Horm. IGF Res. 1999, 9, 174–178. [Google Scholar] [CrossRef]
- Shalita-Chesner, M.; Katz, J.; Shemer, J.; Werner, H. Regulation of insulin-like growth factor-I receptor gene expression by tumor necrosis factor-α and interferon-γ. Mol. Cell. Endocrinol. 2001, 176, 1–12. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Regulation of IGF-1 signaling by microRNAs. Front. Genet. 2015, 5, 472. [Google Scholar] [CrossRef] [PubMed]
- Deming, S.L.; Ren, Z.; Wen, W.; Shu, X.O.; Cai, Q.; Gao, Y.-T.; Zheng, W. Genetic variation in IGF1, IGF-1R, IGFALS, and IGFBP3 in breast cancer survival among Chinese women: A report from the Shanghai Breast Cancer Study. Breast Cancer Res. Treat. 2007, 104, 309–319. [Google Scholar] [CrossRef]
- De Alencar, S.A.; Lopes, J.C.D. A Comprehensive In Silico Analysis of the Functional and Structural Impact of SNPs in theIGF1RGene. J. Biomed. Biotechnol. 2010, 2010, 1–8. [Google Scholar] [CrossRef]
- Kang, H.-S.; Ahn, S.H.; Mishra, S.K.; Hong, K.-M.; Lee, E.S.; Shin, K.H.; Ro, J.; Lee, K.S.; Kim, M.K. Association of Polymorphisms and Haplotypes in the Insulin-Like Growth Factor 1 Receptor (IGF1R) Gene with the Risk of Breast Cancer in Korean Women. PLoS ONE 2014, 9, e84532. [Google Scholar] [CrossRef] [PubMed]
- Gately, K.; Forde, L.; Gray, S.; Morris, D.; Corvin, A.; Tewari, P.; O’Byrne, K. Mutational analysis of the insulin-like growth factor 1 receptor tyrosine kinase domain in non-small cell lung cancer patients. Mol. Clin. Oncol. 2015, 3, 1073–1079. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cho, S.H.; Kim, S.K.; Kwon, E.; Park, H.J.; Kwon, K.H.; Chung, J.-H. Polymorphism ofIGF1RIs Associated with Papillary Thyroid Carcinoma in a Korean Population. J. Interf. Cytokine Res. 2012, 32, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.-A.; Yourk, V.; Farhat, A.; Guo, K.L.; Garcia, A.; Meyskens, F.L.; Liu-Smith, F. A Possible Link of Genetic Variations in ER/IGF1R Pathway and Risk of Melanoma. Int. J. Mol. Sci. 2020, 21, 1776. [Google Scholar] [CrossRef] [PubMed]
- Fragkioudaki, S.; Mavragani, C.P.; Moutsopoulos, H.M. Predicting the risk for lymphoma development in Sjogren syndrome: An Easy Tool for Clinical Use. Medicine 2016, 95, e3766. [Google Scholar] [CrossRef]
- Goules, A.V.; Argyropoulou, O.D.; Pezoulas, V.C.; Chatzis, L.; Critselis, E.; Gandolfo, S.; Ferro, F.; Binutti, M.; Donati, V.; Callegher, S.Z.; et al. Primary Sjögren’s Syndrome of Early and Late Onset: Distinct Clinical Phenotypes and Lymphoma Development. Front. Immunol. 2020, 11, 594096. [Google Scholar] [CrossRef]
- Flores-Chávez, A.; Kostov, B.; Solans, R.; Fraile, G.; Maure, B.; Feijoo-Massó, C.; Rascón, F.-J.; Pérez-Alvarez, R.; Zamora-Pasadas, M.; García-Pérez, A.; et al. Severe, life-threatening phenotype of primary Sjögren’s syndrome: Clinical characterisation and outcomes in 1580 patients (GEAS-SS Registry). Clin. Exp. Rheumatol. 2018, 36, 121–129. [Google Scholar]
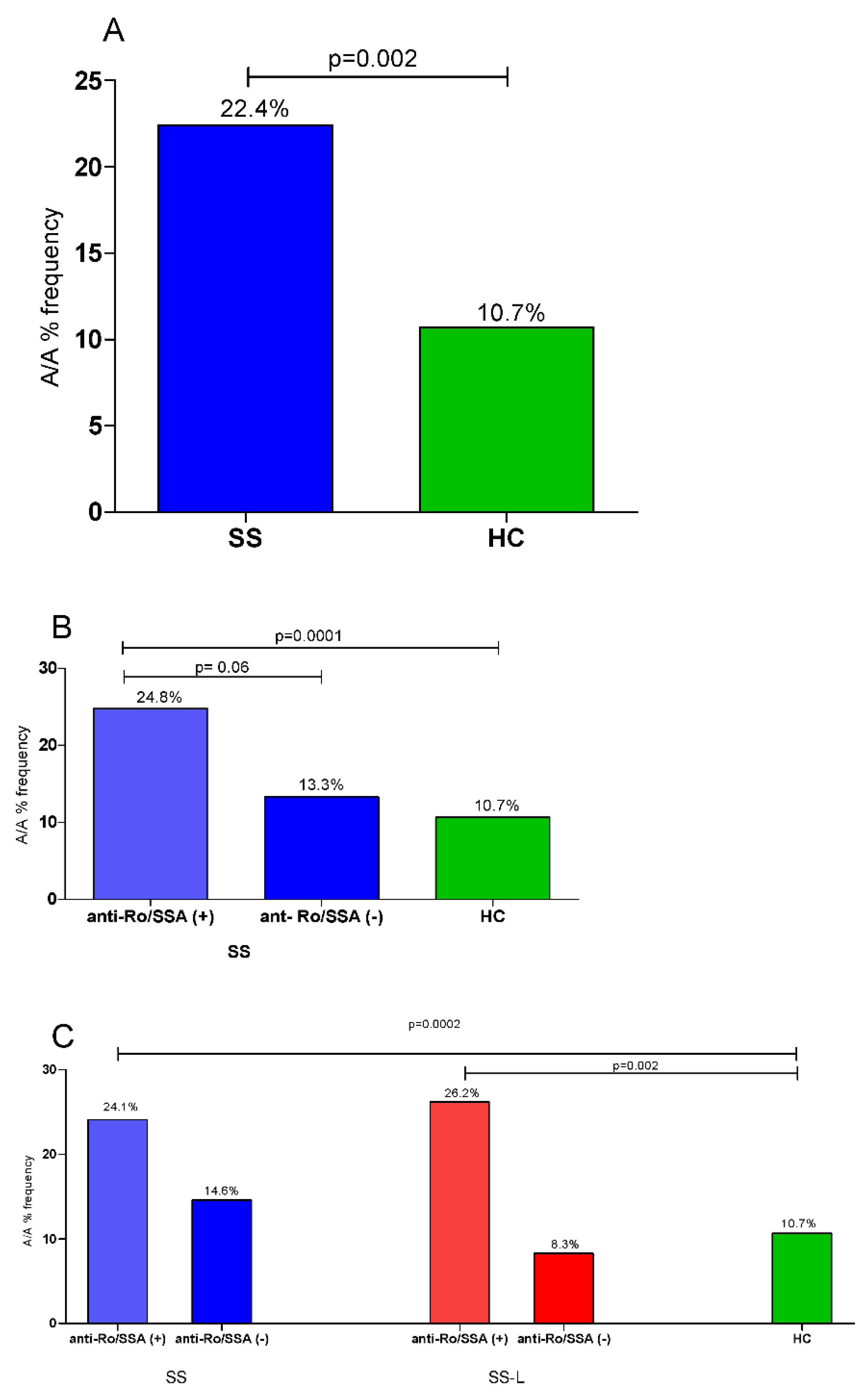

| SNP | Allele | HC (n = 336) (%) | SS (n = 277) (%) | SS-nL (n = 200) (%) | SS-L (n = 77) (%) | * OR [95% CI] p-Value | ** OR [95% CI] p-Value | *** OR [95% CI] p-Value |
|---|---|---|---|---|---|---|---|---|
| rs2229765 | G | 61.3% | 55.4% | 55.5% | 55.2% | 1.27 [1.01–1.60] 0.04 | 1.27 [0.99–1.60] 0.06 | 1.28 [0.90–1.80] 0.16 |
| A | 38.7% | 44.6% | 44.5% | 44.8% |
| SNP | Model | Genotype | HC (n = 337) | SS (n = 277) | SS-nL (n = 200) | SS-L (n = 77) | * OR [95% CI] | ** OR [95% CI] | *** OR [95% CI] | p-Value | p-Value | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2229765 | Codominant | G/G | 33.2 % | 33.2% | 33.2% | 33.8% | 1.00 | 1.00 | 1.00 | 0.005 | 0.012 | 0.27 |
| G/A | 56.1% | 44.4% | 44.4% | 42.9% | 0.81 (0.49–1.32) | 0.80 (0.48–1.48) | 0.92 (0.44–1.95) | |||||
| A/A | 10.7% | 22.4% | 22.4% | 23.4% | 2.30 (1.17–4.52) | 2.42 (1.15–5.12) | 2.11 (0.73–6.08 | |||||
| Dominant | G/G | 33.2% | 33.2% | 33% | 33.8% | 1.00 | 1.00 | 1.00 | 0.83 | 0.7 | 0.8 | |
| G/A-A/A | 66.8% | 66.8% | 67% | 66.2% | 1.05 (0.66–1.67) | 1.11 (0.66–1.89) | 1.10 (0.54–2.23) | |||||
| Recessive | G/G-G/A | 89.3% | 77.6% | 78% | 76.6% | 1.00 | 1.00 | 1.00 | 0.002 | 0.004 | 0.11 | |
| A/A | 10.7% | 22.4% | 22.4% | 23.4% | 2.61 (1.42–4.80) | 2.68 (1.37–5.26) | 2.21 (0.84–5.79) | |||||
| Overdominant | G/G-A/A | 43.9 % | 55.6% | 55% | 57.1% | 1.00 | 1.00 | 1.00 | 0.033 | 0.07 | 0.4 | |
| G/A | 56.1 % | 44.4% | 45% | 42.9% | 0.62 (0.40–0.96) | 0.63 (0.38–1.04) | 0.75 (0.38–1.47 | |||||
| Log-additive | 1.35 (0.98–1.86) | 1.41 (0.98–2.02) | 1.31 (0.78–2.19) | 0.07 | 0.06 | 0.31 |
| Clinical/Serological Characteristics of the SS Cohort | |||
|---|---|---|---|
| Within G/G-G/A Genotype | Within A/A Genotype | p-Values | |
| Demographics | |||
| Mean age (years) (± SD) | 58.4 ± 12.6 | 58.6 ± 15.0 | ns |
| Gender/Female, n (%) | 90.9 | 96.8 | ns |
| Age at SS diagnosis (mean ± SD) | 53.0 ± 13.0 | 49.4 ± 13.6 | 0.035 |
| Clinical features | |||
| Dry mouth, n (%) | 93.9 | 95.2 | ns |
| Dry eyes, n (%) | 88.2 | 90.5 | ns |
| Salivary gland enlargement, n (%) | 30.8 | 43.5 | 0.06 |
| Rose-Bengal stain, n (%) | 57.7 | 54.8 | ns |
| Abnormal Schirmer’s test | 81.3 | 86.4 | ns |
| Arthralgias-Myalgias, n (%) | 64.9 | 68.3 | ns |
| Raynaud’s phenomenon, n (%) | 27.1 | 31.7 | ns |
| Palpable Purpura, n (%) | 15.2 | 15.9 | ns |
| Serological Features | |||
| Anti-Ro/SSA, n (%) | 75.2 | 86.7 | 0.06 |
| Anti-La/SSB, n (%) | 39.5 | 41.7 | ns |
| Rheumatoid Factor positivity, (> 20 IU/mL) (%) | 56.4 | 68.5 | ns |
| WBC (mean ± SD/mm3) | 5633 ± 1967 | 5543 ± 2281 | ns |
| Neutrophils (mean ± SD/mm3) | 3363 ±1711 | 3366 ± 1696 | ns |
| Monocytes (mean ± SD/mm3) | 413 ± 197 | 337 ± 156 | 0.016 |
| Lymphocytes (mean ± SD/mm3) | 1631 ± 687 | 1696 ± 789 | ns |
| HGB g/dL (mean ± SD) | 13 ± 4.08 | 12 ± 1.28 | 0.013 |
| Erythrocyte sedimentation rate SR mm/hour (mean ± SD) | 31.63 ± 24,75 | 35.74 ± 29.32 | ns |
| IgG mg/dL (mean ± SD) | 1687.07 ± 785.86 | 1884.66 ± 1016.98 | ns |
| IgM mg/dL (mean ± SD) | 159.24 ± 125.24 | 236.49 ± 281.73 | ns |
| IgA mg/dL (mean ± SD) | 278.49 ± 118.73 | 337.25 ± 165.56 | ns |
| LDH IU/L (mean ± SD) | 237.91 ± 93.96 | 252.45 ± 111.15 | ns |
| C3 mg/dL (mean ± SD) | 112.29 ± 66.54 | 103.43 ± 25.72 | ns |
| C4 mg/dL (mean ± SD) | 19.16 ± 9.44 | 16.81 ± 7.11 | ns |
| Histopathological features | |||
| Focus score number of lymphocytic infiltrates/4 mm2 (mean ± SD) | 2.41 ± 2.49 | 2.06 ± 1.47 | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skarlis, C.; Marketos, N.; Nezos, A.; Papanikolaou, A.; Voulgarelis, M.; Koutsilieris, M.; Moutsopoulos, H.M.; Mavragani, C.P. +3179G/A Insulin-Like Growth Factor-1 Receptor Polymorphism: A Novel Susceptibility Contributor in Anti-Ro/SSA Positive Patients with Sjögren’s Syndrome: Potential Clinical and Pathogenetic Implications. J. Clin. Med. 2021, 10, 3960. https://doi.org/10.3390/jcm10173960
Skarlis C, Marketos N, Nezos A, Papanikolaou A, Voulgarelis M, Koutsilieris M, Moutsopoulos HM, Mavragani CP. +3179G/A Insulin-Like Growth Factor-1 Receptor Polymorphism: A Novel Susceptibility Contributor in Anti-Ro/SSA Positive Patients with Sjögren’s Syndrome: Potential Clinical and Pathogenetic Implications. Journal of Clinical Medicine. 2021; 10(17):3960. https://doi.org/10.3390/jcm10173960
Chicago/Turabian StyleSkarlis, Charalampos, Nikolaos Marketos, Adrianos Nezos, Asimina Papanikolaou, Michael Voulgarelis, Michael Koutsilieris, Haralampos M. Moutsopoulos, and Clio P. Mavragani. 2021. "+3179G/A Insulin-Like Growth Factor-1 Receptor Polymorphism: A Novel Susceptibility Contributor in Anti-Ro/SSA Positive Patients with Sjögren’s Syndrome: Potential Clinical and Pathogenetic Implications" Journal of Clinical Medicine 10, no. 17: 3960. https://doi.org/10.3390/jcm10173960
APA StyleSkarlis, C., Marketos, N., Nezos, A., Papanikolaou, A., Voulgarelis, M., Koutsilieris, M., Moutsopoulos, H. M., & Mavragani, C. P. (2021). +3179G/A Insulin-Like Growth Factor-1 Receptor Polymorphism: A Novel Susceptibility Contributor in Anti-Ro/SSA Positive Patients with Sjögren’s Syndrome: Potential Clinical and Pathogenetic Implications. Journal of Clinical Medicine, 10(17), 3960. https://doi.org/10.3390/jcm10173960







