Change in the Strategy of Embryo Selection with Time-Lapse System Implementation—Impact on Clinical Pregnancy Rates
Abstract
:1. Introduction
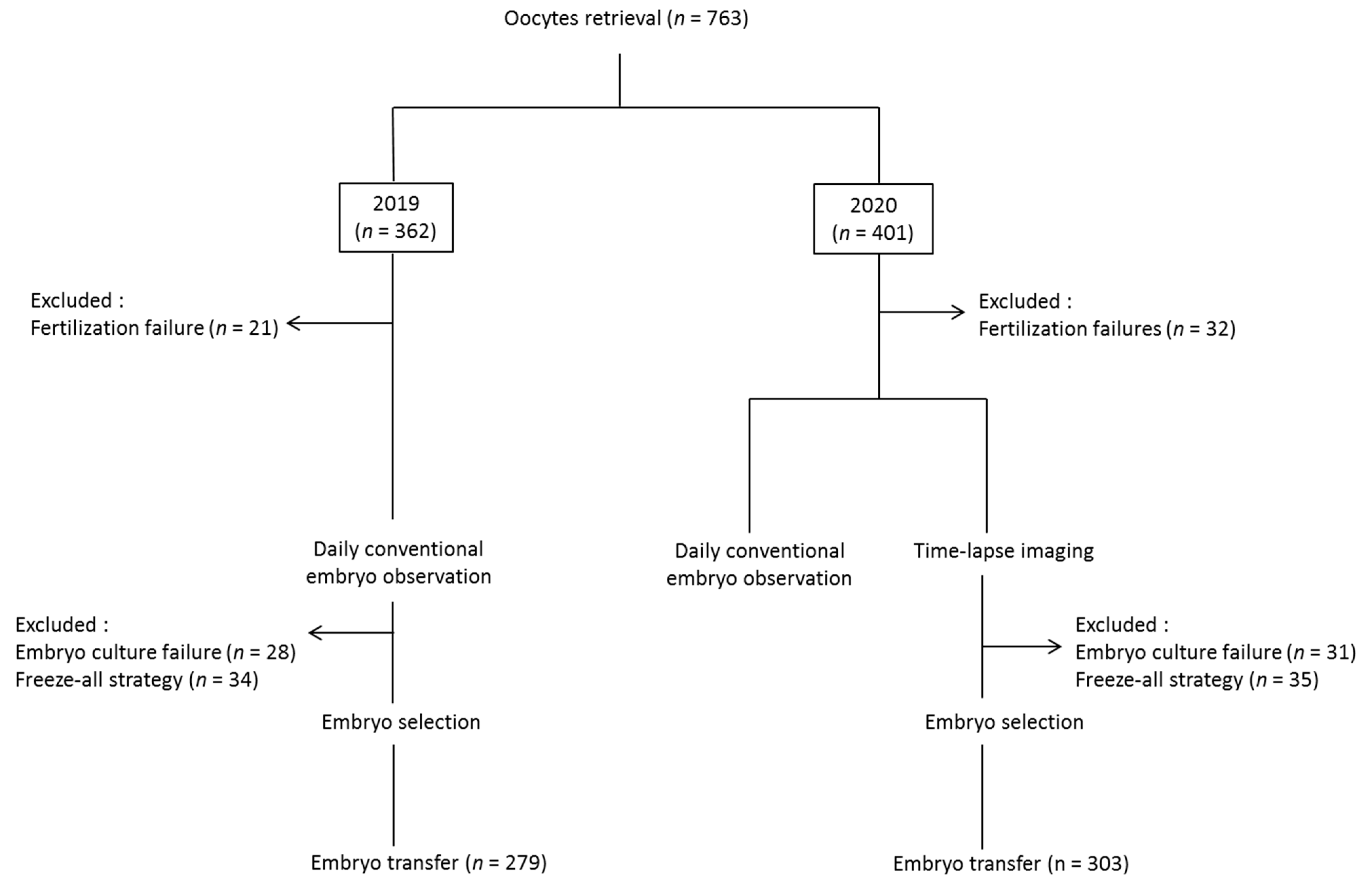
2. Materials and Methods
2.1. Study Design and Population
2.2. Controlled Ovarian Stimulation
2.3. Fertilization and Embryo Culture
2.4. Embryo Transfer and Freezing
2.5. Data Collected
2.6. Outcome Measures
2.7. Statistical Analysis
2.8. Ethical Approval
3. Results
3.1. Daily versus Time-Lapse Observations
3.1.1. Concordance between the Selection Methods
3.1.2. Concordance According to the Day of Transfer
3.1.3. Categories of Morphokinetic Anomalies
3.2. Comparison between TLS and Conventional Incubation and Selection on IVF Outcomes
3.2.1. Patient Characteristics
3.2.2. IVF Parameters and Reproductive Outcomes
3.2.3. Multivariate Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rubio, I.; Kuhlmann, R.; Agerholm, I.; Kirk, J.; Herrero, J.; Escribá, M.-J.; Bellver, J.; Meseguer, M. Limited Implantation Success of Direct-Cleaved Human Zygotes: A Time-Lapse Study. Fertil Steril 2012, 98, 1458–1463. [Google Scholar] [CrossRef]
- Zhan, Q.; Ye, Z.; Clarke, R.; Rosenwaks, Z.; Zaninovic, N. Direct Unequal Cleavages: Embryo Developmental Competence, Genetic Constitution and Clinical Outcome. PLoS ONE 2016, 11, e0166398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-H.; Wu, C.-H.; Chen, Y.-C.; Yang, C.-K.; Wu, T.-H.; Chen, P.-C.; Tsai, H.-D. Effect of Morphokinetics and Morphological Dynamics of Cleavage Stage on Embryo Developmental Potential: A Time-Lapse Study. Taiwan J. Obstet Gynecol 2018, 57, 76–82. [Google Scholar] [CrossRef]
- Desai, N.; Goldberg, J.M.; Austin, C.; Falcone, T. Are Cleavage Anomalies, Multinucleation, or Specific Cell Cycle Kinetics Observed with Time-Lapse Imaging Predictive of Embryo Developmental Capacity or Ploidy? Fertil Steril 2018, 109, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athayde Wirka, K.; Chen, A.A.; Conaghan, J.; Ivani, K.; Gvakharia, M.; Behr, B.; Suraj, V.; Tan, L.; Shen, S. Atypical Embryo Phenotypes Identified by Time-Lapse Microscopy: High Prevalence and Association with Embryo Development. Fertil Steril 2014, 101, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chapple, V.; Roberts, P.; Matson, P. Prevalence, Consequence, and Significance of Reverse Cleavage by Human Embryos Viewed with the Use of the Embryoscope Time-Lapse Video System. Fertil Steril 2014, 102, 1295–1300. [Google Scholar] [CrossRef]
- Barrie, A.; Homburg, R.; McDowell, G.; Brown, J.; Kingsland, C.; Troup, S. Preliminary Investigation of the Prevalence and Implantation Potential of Abnormal Embryonic Phenotypes Assessed Using Time-Lapse Imaging. Reprod Biomed. Online 2017, 34, 455–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, L.R.; Goldberg, J.; Falcone, T.; Austin, C.; Desai, N. Does the Addition of Time-Lapse Morphokinetics in the Selection of Embryos for Transfer Improve Pregnancy Rates? A Randomized Controlled Trial. Fertil Steril 2016, 105, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conaghan, J.; Chen, A.A.; Willman, S.P.; Ivani, K.; Chenette, P.E.; Boostanfar, R.; Baker, V.L.; Adamson, G.D.; Abusief, M.E.; Gvakharia, M.; et al. Improving Embryo Selection Using a Computer-Automated Time-Lapse Image Analysis Test plus Day 3 Morphology: Results from a Prospective Multicenter Trial. Fertil Steril 2013, 100, 412–419. [Google Scholar] [CrossRef]
- Motato, Y.; de los Santos, M.J.; Escriba, M.J.; Ruiz, B.A.; Remohí, J.; Meseguer, M. Morphokinetic Analysis and Embryonic Prediction for Blastocyst Formation through an Integrated Time-Lapse System. Fertil Steril 2016, 105, 376–384. [Google Scholar] [CrossRef]
- Dal Canto, M.; Coticchio, G.; Mignini Renzini, M.; De Ponti, E.; Novara, P.V.; Brambillasca, F.; Comi, R.; Fadini, R. Cleavage Kinetics Analysis of Human Embryos Predicts Development to Blastocyst and Implantation. Reprod Biomed. Online 2012, 25, 474–480. [Google Scholar] [CrossRef]
- Cruz, M.; Garrido, N.; Herrero, J.; Pérez-Cano, I.; Muñoz, M.; Meseguer, M. Timing of Cell Division in Human Cleavage-Stage Embryos Is Linked with Blastocyst Formation and Quality. Reprod Biomed. Online 2012, 25, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Shi, J.X.; Gong, F.; Zhang, S.P.; Lu, C.F.; Tan, K.; Leng, L.Z.; Hao, M.; He, H.; Gu, Y.F.; et al. Cleavage Pattern Predicts Developmental Potential of Day 3 Human Embryos Produced by IVF. Reprod Biomed. Online 2015, 30, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasco, B.; Arroyo, G.; Gil, Y.; Gómez, M.J.; Rodríguez, I.; Barri, P.N.; Veiga, A.; Boada, M. Selecting Embryos with the Highest Implantation Potential Using Data Mining and Decision Tree Based on Classical Embryo Morphology and Morphokinetics. J. Assist. Reprod Genet. 2017, 34, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S.; Reigstad, M.M.; Petersen, B.M.; Schwennicke, A.; Wegner Hausken, J.; Storeng, R. Time-Lapse Imaging Derived Morphokinetic Variables Reveal Association with Implantation and Live Birth Following in Vitro Fertilization: A Retrospective Study Using Data from Transferred Human Embryos. PLoS ONE 2020, 15, e0242377. [Google Scholar] [CrossRef]
- Adolfsson, E.; Porath, S.; Andershed, A.N. External Validation of a Time-Lapse Model; a Retrospective Study Comparing Embryo Evaluation Using a Morphokinetic Model to Standard Morphology with Live Birth as Endpoint. JBRA Assist. Reprod 2018, 22, 205–214. [Google Scholar] [CrossRef]
- Petersen, B.M.; Boel, M.; Montag, M.; Gardner, D.K. Development of a Generally Applicable Morphokinetic Algorithm Capable of Predicting the Implantation Potential of Embryos Transferred on Day 3. Hum. Reprod 2016, 31, 2231–2244. [Google Scholar] [CrossRef] [Green Version]
- Rubio, I.; Galán, A.; Larreategui, Z.; Ayerdi, F.; Bellver, J.; Herrero, J.; Meseguer, M. Clinical Validation of Embryo Culture and Selection by Morphokinetic Analysis: A Randomized, Controlled Trial of the EmbryoScope. Fertil Steril 2014, 102, 1287–1294. [Google Scholar] [CrossRef]
- Meseguer, M.; Herrero, J.; Tejera, A.; Hilligsøe, K.M.; Ramsing, N.B.; Remohí, J. The Use of Morphokinetics as a Predictor of Embryo Implantation. Hum. Reprod 2011, 26, 2658–2671. [Google Scholar] [CrossRef] [Green Version]
- Basile, N.; Vime, P.; Florensa, M.; Aparicio Ruiz, B.; García Velasco, J.A.; Remohí, J.; Meseguer, M. The Use of Morphokinetics as a Predictor of Implantation: A Multicentric Study to Define and Validate an Algorithm for Embryo Selection. Hum. Reprod 2015, 30, 276–283. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chapple, V.; Feenan, K.; Roberts, P.; Matson, P. Time-Lapse Deselection Model for Human Day 3 in Vitro Fertilization Embryos: The Combination of Qualitative and Quantitative Measures of Embryo Growth. Fertil Steril 2016, 105, 656–662. [Google Scholar] [CrossRef] [Green Version]
- Milewski, R.; Ajduk, A. Time-Lapse Imaging of Cleavage Divisions in Embryo Quality Assessment. Reproduction 2017, 154, R37–R53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ESHRE Working group on Time-lapse, technology; Apter, S.; Ebner, T.; Freour, T.; Guns, Y.; Kovacic, B.; Le Clef, N.; Marques, M.; Meseguer, M.; Montjean, D.; et al. Good Practice Recommendations for the Use of Time-Lapse Technology†. Hum. Reprod Open 2020, 2020, hoaa008. [Google Scholar] [CrossRef]
- Storr, A.; Venetis, C.; Cooke, S.; Kilani, S.; Ledger, W. Time-Lapse Algorithms and Morphological Selection of Day-5 Embryos for Transfer: A Preclinical Validation Study. Fertil Steril 2018, 109, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Qi, F.; Matson, P.; Morbeck, D.E.; Mol, B.W.; Zhao, S.; Afnan, M. Between-Laboratory Reproducibility of Time-Lapse Embryo Selection Using Qualitative and Quantitative Parameters: A Systematic Review and Meta-Analysis. J. Assist. Reprod Genet. 2020, 37, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Fréour, T.; Le Fleuter, N.; Lammers, J.; Splingart, C.; Reignier, A.; Barrière, P. External Validation of a Time-Lapse Prediction Model. Fertil Steril 2015, 103, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Barrie, A.; Homburg, R.; McDowell, G.; Brown, J.; Kingsland, C.; Troup, S. Examining the Efficacy of Six Published Time-Lapse Imaging Embryo Selection Algorithms to Predict Implantation to Demonstrate the Need for the Development of Specific, in-House Morphokinetic Selection Algorithms. Fertil Steril 2017, 107, 613–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meseguer, M.; Rubio, I.; Cruz, M.; Basile, N.; Marcos, J.; Requena, A. Embryo Incubation and Selection in a Time-Lapse Monitoring System Improves Pregnancy Outcome Compared with a Standard Incubator: A Retrospective Cohort Study. Fertil Steril 2012, 98, 1481–1489. [Google Scholar] [CrossRef]
- Adamson, G.D.; Abusief, M.E.; Palao, L.; Witmer, J.; Palao, L.M.; Gvakharia, M. Improved Implantation Rates of Day 3 Embryo Transfers with the Use of an Automated Time-Lapse-Enabled Test to Aid in Embryo Selection. Fertil Steril 2016, 105, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Barrie, A.; Homburg, R.; McDowell, G.; Brown, J.; Kingsland, C.; Troup, S. Embryos Cultured in a Time-Lapse System Result in Superior Treatment Outcomes: A Strict Matched Pair Analysis. Hum. Fertil 2017, 20, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Han, W.; Wang, J.; Zhang, X.; Liu, W.; Xiong, S.; Han, S.; Liu, J.; Gao, Y.; Huang, G. Embryo Culture Using a Time-Lapse Monitoring System Improves Live Birth Rates Compared with a Conventional Culture System: A Prospective Cohort Study. Hum. Fertil 2018, 21, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Siristatidis, C.; Komitopoulou, M.A.; Makris, A.; Sialakouma, A.; Botzaki, M.; Mastorakos, G.; Salamalekis, G.; Bettocchi, S.; Palmer, G.A. Morphokinetic Parameters of Early Embryo Development via Time Lapse Monitoring and Their Effect on Embryo Selection and ICSI Outcomes: A Prospective Cohort Study. J. Assist. Reprod Genet. 2015, 32, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pribenszky, C.; Nilselid, A.-M.; Montag, M. Time-Lapse Culture with Morphokinetic Embryo Selection Improves Pregnancy and Live Birth Chances and Reduces Early Pregnancy Loss: A Meta-Analysis. Reprod Biomed. Online 2017, 35, 511–520. [Google Scholar] [CrossRef] [Green Version]
- Magdi, Y.; Samy, A.; Abbas, A.M.; Ibrahim, M.A.; Edris, Y.; El-Gohary, A.; Fathi, A.M.; Fawzy, M. Effect of Embryo Selection Based Morphokinetics on IVF/ICSI Outcomes: Evidence from a Systematic Review and Meta-Analysis of Randomized Controlled Trials. Arch. Gynecol Obstet 2019, 300, 1479–1490. [Google Scholar] [CrossRef]
- Kirkegaard, K.; Hindkjaer, J.J.; Grøndahl, M.L.; Kesmodel, U.S.; Ingerslev, H.J. A Randomized Clinical Trial Comparing Embryo Culture in a Conventional Incubator with a Time-Lapse Incubator. J. Assist. Reprod Genet. 2012, 29, 565–572. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, P.; Matyas, S.; Forgacs, V.; Sajgo, A.; Molnar, L.; Pribenszky, C. Non-Invasive Embryo Evaluation and Selection Using Time-Lapse Monitoring: Results of a Randomized Controlled Study. Eur J. Obstet Gynecol Reprod Biol 2019, 233, 58–63. [Google Scholar] [CrossRef]
- Kahraman, S.; Çetinkaya, M.; Pirkevi, C.; Yelke, H.; Kumtepe, Y. Comparison of Blastocyst Development and Cycle Outcome in Patients with ESET Using Either Conventional or Time Lapse Incubators. A Prospective Study of Good Prognosis Patients. Journal of Reproductive and Stem Cell Biotechnology 2012, 3, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Wei, S.; Hu, J.; Yuan, J.; Liu, F. Does Time-Lapse Imaging Have Favorable Results for Embryo Incubation and Selection Compared with Conventional Methods in Clinical in Vitro Fertilization? A Meta-Analysis and Systematic Review of Randomized Controlled Trials. PLoS ONE 2017, 12, e0178720. [Google Scholar] [CrossRef] [PubMed]
- Reignier, A.; Girard, J.-M.; Lammers, J.; Chtourou, S.; Lefebvre, T.; Barriere, P.; Freour, T. Performance of Day 5 KIDScoreTM Morphokinetic Prediction Models of Implantation and Live Birth after Single Blastocyst Transfer. J. Assist. Reprod Genet. 2019, 36, 2279–2285. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; WHO Press: Geneva, Switzerland, 2010; ISBN 978 92 4 154778 9. [Google Scholar]
- Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul Consensus Workshop on Embryo Assessment: Proceedings of an Expert Meeting. Hum. Reprod. 2011, 26, 1270–1283. [CrossRef] [Green Version]
- Ciray, H.N.; Campbell, A.; Agerholm, I.E.; Aguilar, J.; Chamayou, S.; Esbert, M.; Sayed, S. Time-Lapse User Group Proposed Guidelines on the Nomenclature and Annotation of Dynamic Human Embryo Monitoring by a Time-Lapse User Group. Hum. Reprod 2014, 29, 2650–2660. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2016. Available online: Http://Www.R-Project.Org/ (accessed on 27 June 2021).
- Wong, C.C.; Loewke, K.E.; Bossert, N.L.; Behr, B.; De Jonge, C.J.; Baer, T.M.; Reijo Pera, R.A. Non-Invasive Imaging of Human Embryos before Embryonic Genome Activation Predicts Development to the Blastocyst Stage. Nat. Biotechnol 2010, 28, 1115–1121. [Google Scholar] [CrossRef]
- Ergin, E.G.; Calişkan, E.; Yalçinkaya, E.; Oztel, Z.; Cökelez, K.; Ozay, A.; Ozörnek, H.M. Frequency of Embryo Multinucleation Detected by Time-Lapse System and Its Impact on Pregnancy Outcome. Fertil Steril 2014, 102, 1029–1033. [Google Scholar] [CrossRef]
- Desch, L.; Bruno, C.; Luu, M.; Barberet, J.; Choux, C.; Lamotte, M.; Schmutz, E.; Sagot, P.; Fauque, P. Embryo Multinucleation at the Two-Cell Stage Is an Independent Predictor of Intracytoplasmic Sperm Injection Outcomes. Fertil Steril 2017, 107, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Barberet, J.; Bruno, C.; Valot, E.; Antunes-Nunes, C.; Jonval, L.; Chammas, J.; Choux, C.; Ginod, P.; Sagot, P.; Soudry-Faure, A.; et al. Can Novel Early Non-Invasive Biomarkers of Embryo Quality Be Identified with Time-Lapse Imaging to Predict Live Birth? Hum. Reprod 2019, 34, 1439–1449. [Google Scholar] [CrossRef] [Green Version]
- Marcos, J.; Pérez-Albalá, S.; Mifsud, A.; Molla, M.; Landeras, J.; Meseguer, M. Collapse of Blastocysts Is Strongly Related to Lower Implantation Success: A Time-Lapse Study. Hum. Reprod 2015, 30, 2501–2508. [Google Scholar] [CrossRef]
- Barberet, J.; Chammas, J.; Bruno, C.; Valot, E.; Vuillemin, C.; Jonval, L.; Choux, C.; Sagot, P.; Soudry, A.; Fauque, P. Randomized Controlled Trial Comparing Embryo Culture in Two Incubator Systems: G185 K-System versus EmbryoScope. Fertil Steril 2018, 109, 302–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Bergh, C.; Selleskog, U.; Thurin-Kjellberg, A.; Lundin, K. No Benefit of Culturing Embryos in a Closed System Compared with a Conventional Incubator in Terms of Number of Good Quality Embryos: Results from an RCT. Hum. Reprod 2015, 30, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Kaser, D.J.; Bormann, C.L.; Missmer, S.A.; Farland, L.V.; Ginsburg, E.S.; Racowsky, C. A Pilot Randomized Controlled Trial of Day 3 Single Embryo Transfer with Adjunctive Time-Lapse Selection versus Day 5 Single Embryo Transfer with or without Adjunctive Time-Lapse Selection. Hum. Reprod 2017, 32, 1598–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieslinger, D.C.; De Gheselle, S.; Lambalk, C.B.; De Sutter, P.; Kostelijk, E.H.; Twisk, J.W.R.; van Rijswijk, J.; Van den Abbeel, E.; Vergouw, C.G. Embryo Selection Using Time-Lapse Analysis (Early Embryo Viability Assessment) in Conjunction with Standard Morphology: A Prospective Two-Center Pilot Study. Hum. Reprod 2016, 31, 2450–2457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaninovic, N.; Nohales, M.; Zhan, Q.; de Los Santos, Z.M.J.; Sierra, J.; Rosenwaks, Z.; Meseguer, M. A Comparison of Morphokinetic Markers Predicting Blastocyst Formation and Implantation Potential from Two Large Clinical Data Sets. J. Assist. Reprod Genet. 2019, 36, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Lundin, K.; Park, H. Time-Lapse Technology for Embryo Culture and Selection. Ups J. Med. Sci 2020, 125, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Rocafort, E.; Enciso, M.; Leza, A.; Sarasa, J.; Aizpurua, J. Euploid Embryos Selected by an Automated Time-Lapse System Have Superior SET Outcomes than Selected Solely by Conventional Morphology Assessment. J. Assist. Reprod Genet. 2018, 35, 1573–1583. [Google Scholar] [CrossRef]
- Armstrong, S.; Bhide, P.; Jordan, V.; Pacey, A.; Marjoribanks, J.; Farquhar, C. Time-Lapse Systems for Embryo Incubation and Assessment in Assisted Reproduction. Cochrane Database Syst Rev. 2019, 5, CD011320. [Google Scholar] [CrossRef] [PubMed]
- Polanski, L.T.; Coelho Neto, M.A.; Nastri, C.O.; Navarro, P.A.; Ferriani, R.A.; Raine-Fenning, N.; Martins, W.P. Time-Lapse Embryo Imaging for Improving Reproductive Outcomes: Systematic Review and Meta-Analysis. Ultrasound Obstet Gynecol 2014, 44, 394–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coticchio, G.; Mignini Renzini, M.; Novara, P.V.; Lain, M.; De Ponti, E.; Turchi, D.; Fadini, R.; Dal Canto, M. Focused Time-Lapse Analysis Reveals Novel Aspects of Human Fertilization and Suggests New Parameters of Embryo Viability. Hum. Reprod 2018, 33, 23–31. [Google Scholar] [CrossRef]
- Coticchio, G.; Ezoe, K.; Lagalla, C.; Shimazaki, K.; Ohata, K.; Ninomiya, M.; Wakabayashi, N.; Okimura, T.; Uchiyama, K.; Kato, K.; et al. Perturbations of Morphogenesis at the Compaction Stage Affect Blastocyst Implantation and Live Birth Rates. Hum. Reprod 2021, 36, 918–928. [Google Scholar] [CrossRef]
- Bori, L.; Paya, E.; Alegre, L.; Viloria, T.A.; Remohi, J.A.; Naranjo, V.; Meseguer, M. Novel and Conventional Embryo Parameters as Input Data for Artificial Neural Networks: An Artificial Intelligence Model Applied for Prediction of the Implantation Potential. Fertil Steril 2020, 114, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Barrie, A.; McDowell, G.; Troup, S. An Investigation into the Effect of Potential Confounding Patient and Treatment Parameters on Human Embryo Morphokinetics. Fertility and Sterility 2021, S0015028220325516. [Google Scholar] [CrossRef]
- Khosravi, P.; Kazemi, E.; Zhan, Q.; Malmsten, J.E.; Toschi, M.; Zisimopoulos, P.; Sigaras, A.; Lavery, S.; Cooper, L.A.D.; Hickman, C.; et al. Deep Learning Enables Robust Assessment and Selection of Human Blastocysts after in Vitro Fertilization. NPJ Digit. Med. 2019, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.; Cooke, S.; Illingworth, P.J.; Gardner, D.K. Deep Learning as a Predictive Tool for Fetal Heart Pregnancy Following Time-Lapse Incubation and Blastocyst Transfer. Hum. Reprod 2019, 34, 1011–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Tao, W.; Liu, H.; Yu, G.; Li, M.; Ma, S.; Wu, K. Morphokinetic Parameters from a Time-Lapse Monitoring System Cannot Accurately Predict the Ploidy of Embryos. J. Assist. Reprod Genet. 2017, 34, 1173–1178. [Google Scholar] [CrossRef]
- Rienzi, L.; Capalbo, A.; Stoppa, M.; Romano, S.; Maggiulli, R.; Albricci, L.; Scarica, C.; Farcomeni, A.; Vajta, G.; Ubaldi, F.M. No Evidence of Association between Blastocyst Aneuploidy and Morphokinetic Assessment in a Selected Population of Poor-Prognosis Patients: A Longitudinal Cohort Study. Reprod Biomed. Online 2015, 30, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Minasi, M.G.; Colasante, A.; Riccio, T.; Ruberti, A.; Casciani, V.; Scarselli, F.; Spinella, F.; Fiorentino, F.; Varricchio, M.T.; Greco, E. Correlation between Aneuploidy, Standard Morphology Evaluation and Morphokinetic Development in 1730 Biopsied Blastocysts: A Consecutive Case Series Study. Hum. Reprod 2016, 31, 2245–2254. [Google Scholar] [CrossRef] [Green Version]
- Reignier, A.; Lammers, J.; Barriere, P.; Freour, T. Can Time-Lapse Parameters Predict Embryo Ploidy? A Systematic Review. Reprod Biomed. Online 2018, 36, 380–387. [Google Scholar] [CrossRef] [Green Version]
- Gazzo, E.; Peña, F.; Valdéz, F.; Chung, A.; Bonomini, C.; Ascenzo, M.; Velit, M.; Escudero, E. The KidscoreTM D5 Algorithm as an Additional Tool to Morphological Assessment and PGT-A in Embryo Selection: A Time-Lapse Study. JBRA Assist. Reprod 2020, 24, 55–60. [Google Scholar] [CrossRef]
- Alegre, L.; Del Gallego, R.; Arrones, S.; Hernández, P.; Muñoz, M.; Meseguer, M. Novel Noninvasive Embryo Selection Algorithm Combining Time-Lapse Morphokinetics and Oxidative Status of the Spent Embryo Culture Medium. Fertil Steril 2019, 111, 918–927. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, M.; Liu, K. The Influence of Seasonal Variations on in Vitro Fertilization and Fresh/Frozen Embryo Transfer: A Retrospective Study. Arch. Gynecol Obstet 2018, 298, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, H.; Mol, B.W.; Shi, W.; Gao, M.; Shi, J. Seasonal Variability Does Not Impact in Vitro Fertilization Success. Sci Rep. 2019, 9, 17185. [Google Scholar] [CrossRef] [PubMed]
| n (%) | |
|---|---|
| Direct cleavage | 23 (28%) |
| Reverse cleavage | 4 (4.9%) |
| Multinucleation | 17 (20.7%) |
| Abnormal pronuclei | 1 (1.2%) |
| None | 37 (45.1%) |
| n (%) | |
|---|---|
| Detected abnormal cleavage associated with a decrease in KIDScore™ | 26 (46.4%) |
| Lower KIDScore™ only | 15 (26.8%) |
| Multinucleation | 5 (8.9%) |
| Subjective judgment | 10 (17.9%) |
| Conventional Observation Year 2019 (n = 279) | Time-Lapse Observation Year 2020 (n = 303) | p-Value | |
|---|---|---|---|
| Female age (year) | 33.1 ± 4.6 | 33.7 ± 4.7 | 0.1 |
| BMI (kg/m2) | 24.7 ± 4.9 | 23.8 ± 4.9 | 0.03 |
| Tobacco use | |||
| - Never | 207 (74.2%) | 227 (75.2%) | 0.1 |
| - Current | 48 (17.2%) | 37 (12.3%) | |
| - Former | 24 (8.6%) | 38 (12.6%) | |
| Cause of female infertility | 0.2 | ||
| - None | 128 (45.9%) | 125 (41.3%) | |
| - DOR | 53 (19.0%) | 53 (17.5%) | |
| - PCOS | 34 (12.2%) | 47 (15.5%) | |
| - Endometriosis | 22 (7.9%) | 23 (7.6%) | |
| - Tubal | 18 (6.5%) | 13 (4.3%) | |
| - Mixed | 24 (8.6%) | 42 (13.9%) | |
| AMH (ng/mL) | 3.0 ± 2.8 | 3.1 ± 2.5 | 0.7 |
| Basal FSH (IU/L) | 7.6 ± 2.9 | 7.7 ± 3.1 | 0.9 |
| Basal LH (IU/L) | 5.6 ± 2.6 | 5.8 ± 2.8 | 0.3 |
| Basal E2 (pg/mL) | 43.2 ± 27.5 | 41.7 ± 21.7 | 0.5 |
| AFC | 19.4 ± 12.7 | 20.6 ± 12.1 | 0.2 |
| Semen parameters | 0.4 | ||
| - Normal | 183 (65.6%) | 208 (68.6%) | |
| - TMSC < 5 millions | 96 (34.4%) | 95 (31.4%) | |
| Stimulation Protocol | 0.02 | ||
| - GnRH Antagonist (%) | 253 (90.7%) | 256 (84.5%) | |
| - GnRH Agonist (%) | 26 (9.3%) | 47 (15.5%) | |
| Gonadotropin type | 0.1 | ||
| - FSH | 175 (62.7%) | 208 (68.6%) | |
| - FSH + LH | 104 (37.3%) | 95 (31.4%) | |
| Total dose of FSH (IU) | 2 509 ± 1 102 | 2 215 ± 870 | <0.001 |
| Conventional Observation Year 2019 (n = 279) | Time-Lapse Observation Year 2020 (n = 303) | p-Value | |
|---|---|---|---|
| Insemination method | 0.1 | ||
| - IVF | 122 (43.7%) | 146 (48.2%) | |
| - ICSI | 153 (54.8%) | 147 (48.5%) | |
| - IVF + ICSI | 4 (1.4%) | 10 (3.3%) | |
| Oocytes retrieved | 10.7 ± 5.6 | 11.2 ± 6.3 | 0.3 |
| Embryos obtained | 5.5 ± 3.5 | 5.7 ± 3.7 | 0.4 |
| Embryo transfer | |||
| - Single embryo transfer | 106 (38.0%) | 130 (42.9%) | 0.2 |
| - Double embryo transfer | 173 (62.0%) | 173 (57.1%) | |
| Stage at embryo transfer | |||
| - Day 2 | 170 (60.9%) | 101 (33.3%) | <0.001 |
| - Day 3 | 92 (33.0%) | 157 (51.8%) | |
| - Blastocyst | 17 (6.1%) | 45 (14.9%) | |
| Biological pregnancy | 67 (24.0%) | 104 (34.3%) | 0.006 |
| Implantation | 74 (16.4%) | 110 (23.1%) | 0.01 |
| Clinical Pregnancy | 61 (21.9%) | 98 (32.3%) | 0.005 |
| Single/Twin Pregnancy | 48/13 | 86/12 |
| Characteristic | OR | 95% CI | p-Value |
|---|---|---|---|
| BMI | 0.99 | 0.95, 1.03 | 0.6 |
| Total dose of FSH | 0.99 | 0.99, 1.00 | 0.016 |
| Stimulation Protocol | |||
| GnRH Agonist | 1.0 | — | |
| GnRH Antagonist | 1.32 | 0.72, 2.52 | 0.4 |
| Stage at embryo transfer | |||
| Day 2 | 1.0 | — | |
| Day 3 | 1.13 | 0.74, 1.72 | 0.6 |
| Day 5 | 1.41 | 0.74, 2.62 | 0.3 |
| Embryo selection method | |||
| Conventional morphology (2019) | 1.0 | — | |
| Time-lapse observation (2020) | 1.57 | 1.05, 2.36 | 0.029 |
| OR = Odds Ratio, CI = Confidence Interval | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boucret, L.; Tramon, L.; Saulnier, P.; Ferré-L’Hôtellier, V.; Bouet, P.-E.; May-Panloup, P. Change in the Strategy of Embryo Selection with Time-Lapse System Implementation—Impact on Clinical Pregnancy Rates. J. Clin. Med. 2021, 10, 4111. https://doi.org/10.3390/jcm10184111
Boucret L, Tramon L, Saulnier P, Ferré-L’Hôtellier V, Bouet P-E, May-Panloup P. Change in the Strategy of Embryo Selection with Time-Lapse System Implementation—Impact on Clinical Pregnancy Rates. Journal of Clinical Medicine. 2021; 10(18):4111. https://doi.org/10.3390/jcm10184111
Chicago/Turabian StyleBoucret, Lisa, Léa Tramon, Patrick Saulnier, Véronique Ferré-L’Hôtellier, Pierre-Emmanuel Bouet, and Pascale May-Panloup. 2021. "Change in the Strategy of Embryo Selection with Time-Lapse System Implementation—Impact on Clinical Pregnancy Rates" Journal of Clinical Medicine 10, no. 18: 4111. https://doi.org/10.3390/jcm10184111






