Review: Influence of the CYP450 Genetic Variation on the Treatment of Psychotic Disorders
Abstract
:1. Introduction
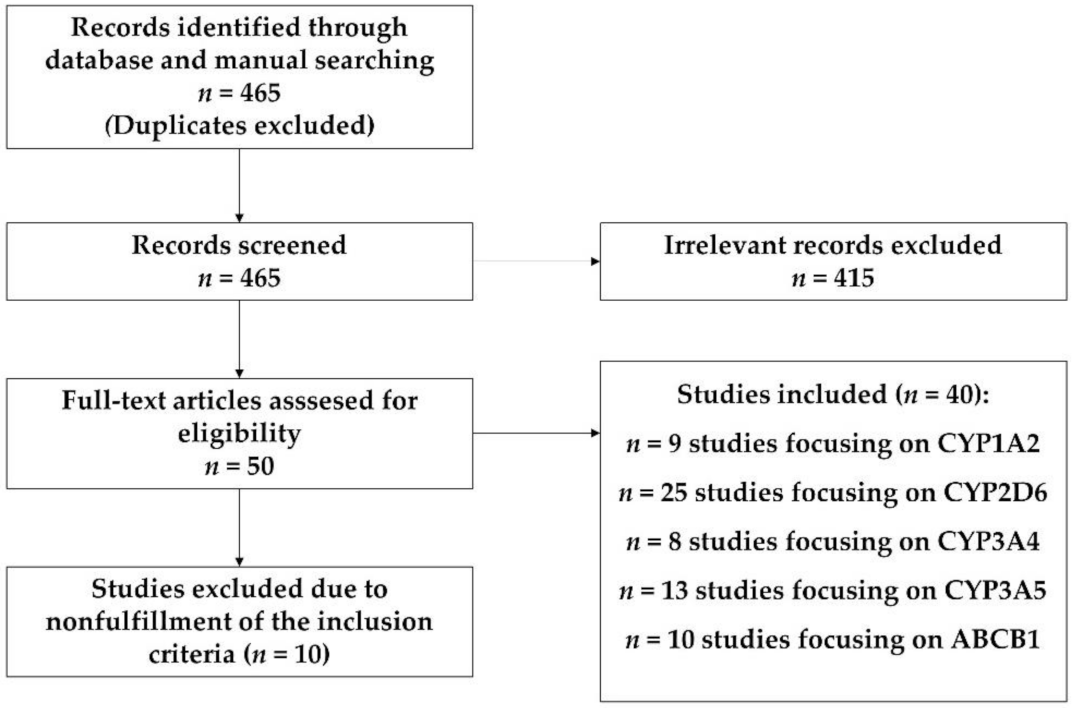
2. Materials and Methods
3. Results
3.1. CYP1A2
3.2. CYP2D6
3.3. CYP3A4
3.4. CYP3A5
3.5. ABCB1
3.6. Summary
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- McGrath, J.; Saha, S.; Chant, D.; Welham, J. Schizophrenia: A Concise Overview of Incidence, Prevalence, and Mortality. Epidemiol. Rev. 2008, 30, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Lally, J.; MacCabe, J.H. Antipsychotic medication in schizophrenia: A review. Br. Med. Bull. 2015, 114, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Ravyn, D.; Ravyn, V.; Lowney, R.; Nasrallah, H.A. CYP450 Pharmacogenetic treatment strategies for antipsychotics: A review of the evidence. Schizophr. Res. 2013, 149, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, Y.; Sukegawa, T.; Inagaki, A.; Inada, T.; Yoshio, T.; Yoshimura, R.; Iwata, N. Evaluation of the individual safe correction of antipsychotic agent polypharmacy in Japanese patients with chronic schizophrenia: Validation of safe corrections for anti-psychotic polypharmacy and the high-dose method. Int. J. Neuropsychopharmacol. 2015, 18, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbild, L.; Andersen, S.E.; Werge, T.; Rasmussen, H.B.; Jürgens, G. Does pharmacogenetic testing for CYP450 2D6 and 2C19 among patients with diagnoses within the schizophrenic spectrum reduce treatment costs? Basic Clin. Pharmacol. Toxicol. 2013, 113, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Kasteridis, P.; Ride, J.; Gutacker, N.; Aylott, L.; Dare, C.; Doran, T.; Gilbody, S.; Goddard, M.; Gravelle, H.; Kendrick, T.; et al. Association Between Antipsychotic Polypharmacy and Outcomes for People with Serious Mental Illness in England. Psychiatr. Serv. 2019, 70, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.K. Antipsychotic Polypharmacy: A Dirty Little Secret or a Fashion? Int. J. Neuropsychopharmacol. 2020, 23, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Kamei, H.; Yamada, H.; Hatano, M.; Hanya, M.; Yamada, S.; Iwata, N. Effectiveness in Switching from Antipsychotic Polypharmacy to Monotherapy in Patients with Schizophrenia: A Case Series. Clin. Psychopharmacol. Neurosci. 2020, 18, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S.; Amrtavarshini, R.; Bhandary, R.P.; Praharaj, S.K. Frequency, reasons, and factors associated with antipsychotic polypharmacy in Schizophrenia: A retrospective chart review in a tertiary hospital in India. Asian J. Psychiatry 2020, 51, 102022. [Google Scholar] [CrossRef]
- Pina-Camacho, L.; Díaz-Caneja, C.M.; Saiz, P.A.; Bobes, J.; Corripio, I.; Grasa, E.; Rodriguez-Jimenez, R.; Fernández, M.; Sanjuán, J.; García-López, A.; et al. Estudio farmacogenético del tratamiento a largo plazo con antipsicóticos de segunda generación y sus efectos adversos metabólicos (Estudio SLiM): Justificación, objetivos, diseño y descripción de la muestra. Rev. Psiquiatr. Salud Ment. 2014, 7, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Cao, T.; Wu, X.; Tang, M.; Xiang, D.; Cai, H. Progress in genetic polymorphisms related to lipid disturbances induced by atypical antipsychotic drugs. Front. Pharmacol. 2020, 10, 1669. [Google Scholar] [CrossRef] [Green Version]
- Liperoti, R.; Bernabei, R.; Onder, G. Managing Antipsychotic Medications in Schizophrenia: Comprehensive Assessment and Personalized Care to Improve Clinical Outcomes and Reduce Costs. J. Clin. Psychiatry 2015, 76, e1159–e1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.S.; McIntyre, R.S.; Gentle, J.E.; Park, N.S.; Chiriboga, D.A.; Lee, Y.; Singh, S.; McPherson, M.A. A computational algorithm for personalized medicine in schizophrenia. Schizophr. Res. 2017, 192, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Osmanova, D.Z.; Freidin, M.B.; Fedorenko, O.Y.; Pozhidaev, I.V.; Boiko, A.S.; Vyalova, N.M.; Tiguntsev, V.V.; Kornetova, E.G.; Loonen, A.J.M.; Semke, A.V.; et al. A pharmacogenetic study of patients with schizophrenia from West Siberia gets insight into dopaminergic mechanisms of antipsychotic-induced hy-perprolactinemia. BMC Med. Genet. 2019, 20 (Suppl. 1), 47. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.-F.; Goldstein, D.B.; Angrist, M.; Cavalleri, G. Personalized Medicine and Human Genetic Diversity. Cold Spring Harb. Perspect. Med. 2014, 4, a008581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef] [Green Version]
- Beckmann, J.S.; Lew, D. Reconciling evidence-based medicine and precision medicine in the era of big data: Challenges and opportunities. Genome Med. 2016, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Ramiro, F.; Peiró-Pastor, R.; Aguado, B. Human genomics projects and precision medicine. Gene Ther. 2017, 24, 551–561. [Google Scholar] [CrossRef]
- Moons, T.; De Roo, M.; Claes, S.; Dom, G. Relationship between P-glycoprotein and second-generation antipsychotics. Pharmacogenomics 2011, 12, 1193–1211. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting sys-tematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [Green Version]
- Novalbos, J.; López-Rodríguez, R.; Román, M.; Gallego-Sandín, S.; Ochoa, D.; Abad-Santos, F. Effects of CYP2D6 Genotype on the Pharmacokinetics, Pharmacodynamics, and Safety of Risperidone in Healthy Volunteers. J. Clin. Psychopharmacol. 2010, 30, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Zhao, X.; Zhou, Y.; Duan, J.L.; Cui, Y.M. Effect of CYP2D6, CYP3A5, and MDR1 genetic polymorphisms on the phar-macokinetics of risperidone and its active moiety. J. Clin. Pharmacol. 2010, 50, 659–666. [Google Scholar] [CrossRef]
- Yoo, H.-D.; Cho, H.-Y.; Lee, S.-N.; Yoon, H.; Lee, Y.-B. Population pharmacokinetic analysis of risperidone and 9-hydroxyrisperidone with genetic polymorphisms of CYP2D6 and ABCB1. J. Pharmacokinet. Pharmacodyn. 2012, 39, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-A.; Joo, H.-J.; Lee, H.-M.; Park, J.-Y. Influence of ABCB1 and CYP3A5 genetic polymorphisms on the pharmacokinetics of quetiapine in healthy volunteers. Pharmacogenet. Genom. 2014, 24, 35–42. [Google Scholar] [CrossRef]
- Shilbayeh, S.A.R.; Sy, S.K.B.; Melhem, M.; Zmeili, R.; Derendorf, H. Quantitation of the impact ofCYP3A5A6986G polymorphism on quetiapine pharmacokinetics by simulation of target attainment. Clin. Pharmacol. Drug Dev. 2015, 4, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, T.; López-Rodríguez, R.; Roman, M.; Ochoa, D.; Novalbos, J.; Borobia, A.; Carcas, A.; Abad-Santos, F. Pharmacogenetics of quetiapine in healthy volunteers. Int. Clin. Psychopharmacol. 2015, 30, 82–88. [Google Scholar] [CrossRef]
- Cabaleiro, T.; Ochoa, D.; Román, M.; Moreno, I.; López-Rodríguez, R.; Novalbos, J.; Abad-Santos, F. Polymorphisms inCYP2D6have a Greater Effect on Variability of Risperidone Pharmacokinetics than Gender. Basic Clin. Pharmacol. Toxicol. 2014, 116, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, C.; Ochoa, D.; Roman, M.; Saiz-Rodríguez, M.; Wojnicz, A.; Gómez-Sánchez, C.I.; Martin-Vilchez, S.; Abad-Santos, F. Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 2018, 122, 596–605. [Google Scholar] [CrossRef] [Green Version]
- CYP1A2 Cytochrome P450 Family 1 Subfamily A Member 2 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/1544 (accessed on 28 July 2021).
- Hoffmann, M.F.; Preissner, S.C.; Nickel, J.; Dunkel, M.; Preissner, R.; Preissner, S. The Transformer database: Biotransformation of xenobiotics. Nucleic Acids Res. 2013, 42, D1113–D1117. [Google Scholar] [CrossRef] [Green Version]
- Sachse, C.; Brockmöller, J.; Bauer, S.; Roots, I. Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br. J. Clin. Pharmacol. 1999, 47, 445–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chida, M.; Yokoi, T.; Fukui, T.; Kinoshita, M.; Yokota, J.; Kamataki, T. Detection of Three Genetic Polymorphisms in the 5′-Flanking Region and Intron 1 of Human CYP1A2 in the Japanese Population. Jpn. J. Cancer Res. 1999, 90, 899–902. [Google Scholar] [CrossRef]
- Han, X.-M.; Ouyang, D.-S.; Chen, X.-P.; Shu, Y.; Jiang, C.-H.; Tan, Z.-R.; Zhou, H.-H. Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br. J. Clin. Pharmacol. 2002, 54, 540–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, S.C. CYP1A21*F contains the −163C>A substitution and is highly inducible. Pharmacogenet. Genom. 2013, 23, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Balibey, H.; Basoglu, C.; Lundgren, S.; Babaoğlu, M.O.; Yasar, U.; Herken, H.; Rane, A.; Bozkurt, A.; Cetin, M. CYP1A2*1F Polymorphism Decreases Clinical Response to Clozapine in Patients with Schizophrenia. Klin. Psikofarmakol. Bülteni-Bull. Clin. Psychopharmacol. 2011, 21, 93–99. [Google Scholar] [CrossRef]
- Laika, B.; Leucht, S.; Heres, S.; Schneider, H.; Steimer, W. Pharmacogenetics and olanzapine treatment: CYP1A2*1F and sero-tonergic polymorphisms influence therapeutic outcome. Pharmacogenom. J. 2010, 10, 20–29. [Google Scholar] [CrossRef]
- Czerwensky, F.; Leucht, S.; Steimer, W. CYP1A2∗1D and ∗ 1F polymorphisms have a significant impact on olanzapine serum concentrations. Ther. Drug Monit. 2015, 37, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Lua, A.C.; Wu, L.S.-H.; Wu, B.-J.; Lee, S.-M.; Liu, C.-Z. Cigarette smoking has a differential effect on the plasma level of clozapine in Taiwanese schizophrenic patients associated with the CYP1A2 gene −163A/C single nucleotide polymorphism. Psychiatr. Genet. 2016, 26, 172–177. [Google Scholar] [CrossRef]
- Viikki, M.; Kampman, O.; Seppälä, N.; Mononen, N.; Lehtimäki, T.; Leinonen, E. CYP1A2 polymorphism −1545C > T (rs2470890) is associated with increased side effects to clozapine. BMC Psychiatry 2014, 14, 50. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, S.A.; Filipenko, M.L.; Vyalova, N.M.; Voronina, E.N.; Pozhidaev, I.V.; Osmanova, D.Z.; Ivanov, M.V.; Fedorenko, O.Y.; Semke, A.V.; Bokhan, N. CYP1A2 and CYP2D6 Gene Polymorphisms in Schizophrenic Patients with Neuroleptic Drug-Induced Side Effects. Bull. Exp. Biol. Med. 2016, 160, 687–690. [Google Scholar] [CrossRef]
- Yan, P.; Song, M.; Gao, B.; Wang, S.; Wang, S.; Li, J.; Fang, H.; Wang, C.; Shi, J. Association of the genetic polymorphisms of metabolizing enzymes, transporters, target receptors and their interactions with treatment response to olanzapine in chinese han schizophrenia patients. Psychiatry Res. 2020, 293, 113470. [Google Scholar] [CrossRef]
- Söderberg, M.M.; Haslemo, T.; Molden, E.; Dahl, M.-L. Influence of CYP1A1/CYP1A2 and AHR polymorphisms on systemic olanzapine exposure. Pharmacogenet. Genom. 2013, 23, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Yokoi, T.; Mizutani, M.; Kinoshitah, M.; Funayama, M.; Kamataki, T. Genetic Polymorphism in the 5’-Flanking Region of HumanCYP1A2 Gene: Effect on the CYP1A2 Inducibility in Humans. J. Biochem. 1999, 125, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Suda, A.; Miyauchi, M.; Shiraishi, Y.; Saeki, T.; Fukushima, T.; Fujibayashi, M.; Tsujita, N.; Ishii, C.; Ishii, N.; et al. The association of genetic polymorphisms in CYP1A2, UGT1A4, and ABCB1 with autonomic nervous system dysfunction in schizophrenia patients treated with olanzapine. BMC Psychiatry 2020, 20, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CYP2D6 Cytochrome P450 Family 2 Subfamily D Member 6 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/1565 (accessed on 28 July 2021).
- Gaedigk, A.; Simon, S.; Pearce, R.; Bradford, L.; Kennedy, M.; Leeder, J. The CYP2D6 Activity Score: Translating Genotype Information into a Qualitative Measure of Phenotype. Clin. Pharmacol. Ther. 2007, 83, 234–242. [Google Scholar] [CrossRef]
- Nagai, G.; Mihara, K.; Nakamura, A.; Suzuki, T.; Nemoto, K.; Kagawa, S.; Ohta, I.; Arakaki, H.; Kondo, T. Prolactin concentrations during aripiprazole treatment in relation to sex, plasma drugs concentrations and genetic polymorphisms of dopamine D2 receptor and cyto-chrome P450 2D6 in Japanese patients with schizophrenia. Psychiatry Clin. Neurosci. 2012, 66, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, K.; Mihara, K.; Nakamura, A.; Nagai, G.; Kagawa, S.; Suzuki, T.; Kondo, T. Effects of escitalopram on plasma concentrations of aripiprazole and its active metabolite, dehydroaripiprazole, in Japanese patients. Pharmacopsychiatry 2014, 47, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Lisbeth, P.; Vincent, H.; Kristof, M.; Bernard, S.; Manuel, M.; Hugo, N. Genotype and co-medication dependent CYP2D6 metabolic activity: Effects on serum concentrations of aripiprazole, haloperidol, risperidone, paliperidone and zuclopenthixol. Eur. J. Clin. Pharmacol. 2015, 72, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Jukic, M.M.; Smith, R.L.; Haslemo, T.; Molden, E.; Ingelman-Sundberg, M. Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: A retrospective, cohort study. Lancet Psychiatry 2019, 6, 418–426. [Google Scholar] [CrossRef]
- Kneller, L.A.; Abad-Santos, F.; Hempel, G. Physiologically Based Pharmacokinetic Modelling to Describe the Pharmacokinetics of Risperidone and 9-Hydroxyrisperidone According to Cytochrome P450 2D6 Phenotypes. Clin. Pharmacokinet. 2019, 59, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Van der Weide, K.; van der Weide, J. The Influence of the CYP3A4*22 Polymorphism and CYP2D6 Polymorphisms on Serum Concentrations of Aripiprazole, Haloperidol, Pimozide, and Risperidone in Psychiatric Patients. J. Clin. Psychopharmacol. 2015, 35, 228–236. [Google Scholar] [CrossRef]
- Suzuki, Y.; Fukui, N.; Tsuneyama, N.; Watanabe, J.; Ono, S.; Sugai, T.; Saito, M.; Inoue, Y.; Someya, T. Effect of the cytochrome P450 2D6*10 allele on risper-idone metabolism in Japanese psychiatric patients. Hum. Psychopharmacol. 2012, 27, 43–46. [Google Scholar] [CrossRef]
- Bakken, G.V.; Molden, E.; Hermann, M. Impact of genetic variability in CYP2D6, CYP3A5, and ABCB1 on serum concentrations of quetiapine and N-desalkylquetiapine in psychiatric patients. Ther. Drug Monit. 2015, 37, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Koller, D.; Belmonte, C.; Saiz-Rodríguez, M.; Zubiaur, P.; Román, M.; Ochoa, D.; Abad-Santos, F. Effects of aripiprazole on circadian prolactin secretion related to pharmacogenetics in healthy volunteers. Basic Clin. Pharmacol. Toxicol. 2019, 126, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Hendset, M.; Molden, E.; Knape, M.; Hermann, M. Serum Concentrations of Risperidone and Aripiprazole in Subgroups Encoding CYP2D6 Intermediate Metabolizer Phenotype. Ther. Drug Monit. 2014, 36, 80–85. [Google Scholar] [CrossRef]
- CYP3A4 Cytochrome P450 Family 3 Subfamily A Member 4 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/1576 (accessed on 28 July 2021).
- Fukushima-Uesaka, H.; Saito, Y.; Watanabe, H.; Shiseki, K.; Saeki, M.; Nakamura, T.; Kurose, K.; Sai, K.; Komamura, K.; Ueno, K.; et al. Haplotypes ofCYP3A4 and their close linkage withCYP3A5 haplotypes in a Japanese population. Hum. Mutat. 2003, 23, 100. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, A.; Wang, L.; Xuan, J.; Yu, L.; Che, R.; Li, X.; Gu, N.; Lin, Z.; Feng, G.; et al. Relationship between response to risperidone, plasma concentrations of risperidone and CYP3A4 polymorphisms in schizophrenia patients. J. Psychopharmacol. 2009, 24, 1115–1120. [Google Scholar] [CrossRef]
- Wang, D.; Guo, Y.; Wrighton, S.A.; Cooke, G.E.; Sadee, W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenom. J. 2010, 11, 274–286. [Google Scholar] [CrossRef] [Green Version]
- Elens, L.; van Schaik, R.H.; Panin, N.; de Meyer, M.; Wallemacq, P.; Lison, D.; Mourad, M.; Haufroid, V. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics 2011, 12, 1383–1396. [Google Scholar] [CrossRef]
- Elens, L.; Becker, M.L.; Haufroid, V.; Hofman, A.; Visser, L.E.; Uitterlinden, A.G.; Stricker, B.C.; van Schaik, R.H. Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in The Rotterdam Study. Pharmacogenet. Genom. 2011, 21, 861–866. [Google Scholar] [CrossRef]
- Van der Weide, K.; van der Weide, J. The Influence of the CYP3A4*22 Polymorphism on Serum Concentration of Quetiapine in Psychiatric Patients. J. Clin. Psychopharmacol. 2014, 34, 256–260. [Google Scholar] [CrossRef]
- Dienstmann, R.; Patnaik, A.; Garcia-Carbonero, R.; Cervantes, A.; Benavent, M.; Roselló, S.; Tops, B.; Van Der Post, R.S.; Argilés, G.; Skartved, N.J.; et al. Safety and Activity of the First-in-Class Sym004 Anti-EGFR Antibody Mixture in Patients with Refractory Colorectal Cancer. Cancer Discov. 2015, 5, 598–609. [Google Scholar] [CrossRef] [Green Version]
- Kiss, Á.; Menus, Á.; Tóth, K.; Déri, M.; Sirok, D.; Gabri, E.; Belic, A.; Csukly, G.; Bitter, I.; Monostory, K. Phenoconversion of CYP2D6 by inhibitors modifies aripiprazole exposure. Eur. Arch. Psychiatry Clin. Neurosci. 2020, 270, 71–82. [Google Scholar] [CrossRef] [Green Version]
- CYP3A5 Cytochrome P450 Family 3 Subfamily A Member 5 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/1577 (accessed on 28 July 2021).
- Kuehl, P.M.; Zhang, J.; Lin, Y.; Lamba, J.K.; Assem, M.; Schuetz, J.D.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.; Balleine, R.L.; Collins, M.; Liddle, C.; Clarke, C.L.; Gurney, H. CYP3A5 genotype and midazolam clearance in Australian patients receiving chemotherapy*1. Clin. Pharmacol. Ther. 2004, 75, 529–538. [Google Scholar] [CrossRef]
- Haufroid, V.; Mourad, M.; Van Kerckhove, V.; Wawrzyniak, J.; De Meyer, M.; Eddour, D.C.; Malaise, J.; Lison, D.; Squifflet, J.-P.; Wallemacq, P. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal trans-plant patients. Pharmacogenetics 2004, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Josephson, F.; Allqvist, A.; Janabi, M.; Sayi, J.; Aklillu, E.; Jande, M.; Mahindi, M.; Burhenne, J.; Bottiger, Y.; Gustafsson, L.L.; et al. CYP3A5 Genotype has an Impact on the Metabolism of the HIV Protease Inhibitor Saquinavir. Clin. Pharmacol. Ther. 2007, 81, 708–712. [Google Scholar] [CrossRef]
- Suzuki, Y.; Tsuneyama, N.; Fukui, N.; Sugai, T.; Watanabe, J.; Ono, S.; Saito, M.; Someya, T. Impact of the ABCB1 Gene Polymorphism on Plasma 9-Hydroxyrisperidone and Active Moiety Levels in Japanese Patients with Schizophrenia. J. Clin. Psychopharmacol. 2013, 33, 411–414. [Google Scholar] [CrossRef] [PubMed]
- ABCB1 ATP Binding Cassette Subfamily B Member 1 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/5243 (accessed on 28 July 2021).
- Hoffmeyer, S.; Burk, O.; von Richter, O.; Arnold, H.P.; Brockmöller, J.; Johne, A.; Cascorbi, I.; Gerloff, T.; Roots, I.; Eichelbaum, M.; et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 3473–3478. [Google Scholar] [CrossRef] [PubMed]
- Soranzo, N.; Cavalleri, G.L.; Weale, M.E.; Wood, N.W.; Depondt, C.; Marguerie, R.; Sisodiya, S.M.; Goldstein, D.B. Identifying Candidate Causal Variants Responsible for Altered Activity of the ABCB1 Multidrug Resistance Gene. Genome Res. 2004, 14, 1333–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na Takuathung, M.; Hanprasertpong, N.; Teekachunhatean, S.; Koonrungsesomboon, N. Impact of CYP1A2 genetic polymor-phisms on pharmacokinetics of antipsychotic drugs: A systematic review and meta-analysis. Acta Psychiatrica. Scand. 2019, 139, 15–25. [Google Scholar] [CrossRef]
- Lee, S.T.; Ryu, S.; Kim, S.R.; Kim, M.J.; Kim, S.; Kim, J.W.; Lee, S.Y.; Hong, K.S. Association study of 27 annotated genes for clozapine pharmaco-genetics: Validation of preexisting studies and identification of a new candidate gene, ABCB1, for treatment response. J. Clin. Psychopharmacol. 2012, 32, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Krivoy, A.; Gaughran, F.; Weizman, A.; Breen, G.; MacCabe, J.H. Gene polymorphisms potentially related to the pharmacokinetics of clozapine: A systematic review. Int. Clin. Psychopharmacol. 2016, 31, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Ingelman-Sundberg, M.; Sim, S.C.; Gomez, A.; Rodriguez-Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007, 116, 496–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Brown, S.J.; Shan, Y.; Lee, A.M.; Allen, J.D.; Eum, S.; de Leon, J.; Bishop, J.R. CYP2D6 Genetic Polymorphisms and Risperidone Pharmaco-kinetics: A Systematic Review and Meta-analysis. Pharmacotherapy 2020, 40, 632–647. [Google Scholar] [CrossRef]
| Principal Metabolizer | Secondary Metabolizer | Minor Metabolizer | Product | |
|---|---|---|---|---|
| Olanzapine ¶ | CYP1A2 | CYP2C8 | CYP2D6 *,† | Inactive metabolite |
| Aripiprazole | CYP2D6 †,§, CYP3A4 | CYP3A5 | Active metabolite | |
| Risperidone | CYP2D6 † | CYP3A4 † | Active metabolite (paliperidone) | |
| Amisulpride | NO CYP | |||
| Clozapine | CYP1A2 †, CYP3A4 †,§ | CYP2C19 †, CYP2C9 †, CYP2D6 † | CYP3A5 † | Active metabolite, inactive metabolite (CYP1A2/CYP3A4) |
| Paliperidone | NO CYP | |||
| Quetiapine | CYP3A4 | CYP3A5, CYP2D6 *,† | Inactive metabolite | |
| Asenapine | CYP1A2 | CYP2D6 ‡ | Inactive metabolite | |
| Levomepromazine | CYP3A4 † | CYP1A2 | Inactive metabolite | |
| Reference | SNP(s) | Antipsychotic | Influence on Pharmacokinetic Parameters |
|---|---|---|---|
| Laika et al., 2009 | CYP1A2: rs762551 | Olanzapine | ↓PL |
| Söderberg et al., 2013 | CYP1A2: rs762551, rs2472304/AHR: rs4410790, rs4410790/CYP1A1: rs2470893, rs2472297 | Olanzapine | ↓C/D ↑M/D |
| Czerwesnky et al., 2015 | CYP1A2: rs762551, rs35694136 | Olanzapine | rs762551: ↓PL; rs35694136: ↑PL |
| Vikki et al., 2014 | CYP1A2: rs2470890 | Clozapine | ↓PL |
| Balibey et al., 2011 | CYP1A2: rs762551 | Clozapine | ↓RR |
| Huang et al., 2016 | CYP1A2: rs762551 | Clozapine | ↓PL |
| Cabaleiro et al., 2015 | CYP1A2: rs2069514 | Quetiapine | ↑AUC |
| Yan et al., 2020 | CYP1A2: rs2069514 | Olanzapine | None |
| Hattori et al., 2020 | CYP1A2: rs2069514 | Olanzapine | ↓C/D |
| Novalbos et al., 2010 | CYP2D6 (PM) | Risperidone | ↑PL |
| Nagai et al., 2013 | Aripiprazole | ↑PL | |
| Suzuki et al., 2014 | Ariprazole | ↑PL | |
| Bakken et al., 2015 | Quetiapine | ↑PL | |
| Van Der Weide et al., 2015 | Ariprazole/risperidone | ↓AD | |
| Lisbeth et al., 2015 | Risperidone | ↑PL | |
| Belmonte et al., 2017 | Aripiprazole | ↑PL/↓AD | |
| Jukic et al., 2019 | Aripiprazole | ↑PL | |
| Koller et al., 2020 | CYP2D6 | Aripiprazole | ↑Prolactine (PL, AUC, Cmax) |
| Du et al., 2009 | CYP3A4: rs2242480 | Risperidone | ↓PANSS |
| Van der Weide et al., 2014 | CYP3A4: rs35599367 | Quetiapine | ↑PL |
| Van der Weide et al., 2015 | CYP3A4: rs35599367 | Aripiprazole/risperidone | ↑PL |
| Belmonte et al., 2015 | CYP3A5: rs776746 | Aripiprazole | ↓ M/D |
| Kim et al., 2014 | CYP3A5: rs776746 | Quetiapine | ↑PL |
| Suzuki et al., 2014 | ABCB1: rs1045642 | Risperidone | ↑PL |
| Belmonte et al., 2017 | ABCB1: rs1128503 | Aripiprazole | ↑PL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrascal-Laso, L.; Isidoro-García, M.; Ramos-Gallego, I.; Franco-Martín, M.A. Review: Influence of the CYP450 Genetic Variation on the Treatment of Psychotic Disorders. J. Clin. Med. 2021, 10, 4275. https://doi.org/10.3390/jcm10184275
Carrascal-Laso L, Isidoro-García M, Ramos-Gallego I, Franco-Martín MA. Review: Influence of the CYP450 Genetic Variation on the Treatment of Psychotic Disorders. Journal of Clinical Medicine. 2021; 10(18):4275. https://doi.org/10.3390/jcm10184275
Chicago/Turabian StyleCarrascal-Laso, Lorena, María Isidoro-García, Ignacio Ramos-Gallego, and Manuel A. Franco-Martín. 2021. "Review: Influence of the CYP450 Genetic Variation on the Treatment of Psychotic Disorders" Journal of Clinical Medicine 10, no. 18: 4275. https://doi.org/10.3390/jcm10184275
APA StyleCarrascal-Laso, L., Isidoro-García, M., Ramos-Gallego, I., & Franco-Martín, M. A. (2021). Review: Influence of the CYP450 Genetic Variation on the Treatment of Psychotic Disorders. Journal of Clinical Medicine, 10(18), 4275. https://doi.org/10.3390/jcm10184275








