Hodgkin Lymphoma: A Special Microenvironment
Abstract
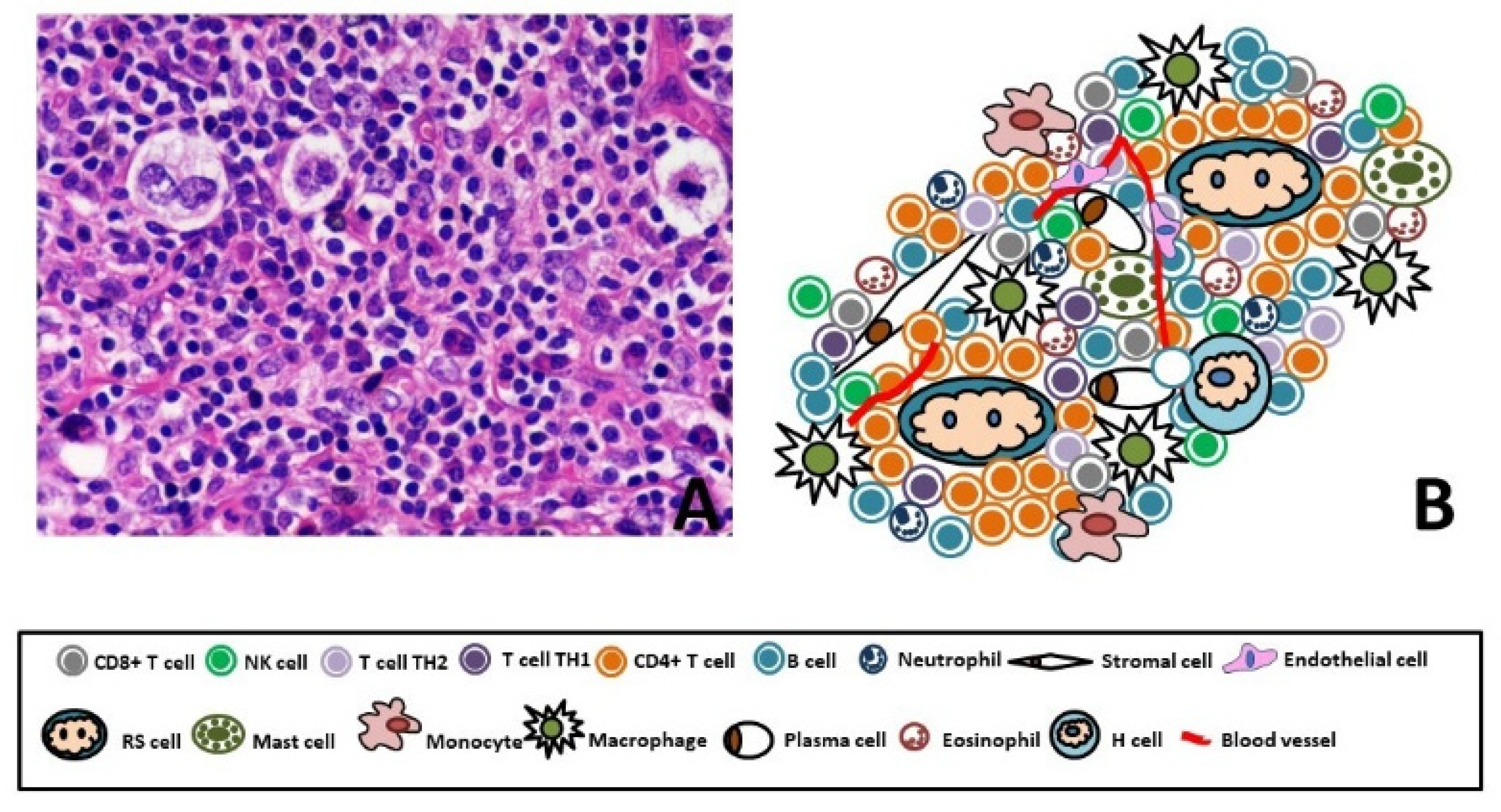
1. Classical Hodgkin’s Lymphoma
2. The Main Component of Tumor Microenvironment (TME)
2.1. T Cell Subsets
2.2. B Lymphocytes
2.3. Macrophages
2.4. Mast Cells
3. cHL and Immunoescape
4. GEP Signature and TME

5. New TME Based Therapeutic Strategy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; Volume 2, ISBN 9789283244943. [Google Scholar]
- Newcom, S.R.; Kadin, M.E.; Phillips, C. L-428 Reed-Sternberg cells and mononuclear Hodgkin’s cells arise from a single cloned mononuclear cell. Int. J. Cell Cloning 1988, 6, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Rengstl, B.; Newrzela, S.; Heinrich, T.; Weiser, C.; Thalheimer, F.B.; Schmid, F.; Warner, K.; Hartmann, S.; Schroeder, T.; Küppers, R.; et al. Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20729–20734. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R.; Schwering, I.; Bräuninger, A.; Rajewsky, K.; Hansmann, M.L. Biology of Hodgkin’s lymphoma. Ann. Oncol. 2002, 13 (Suppl. 1), 11–18. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.S.; Harris, N.L.; Stein, H.; Vardiman, J. Tumours of Haematopoietic and Lymphoid Tissues, 3rd ed.; IARC Press: Lyon, France, 2001. [Google Scholar]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2008; Volume 4, ISBN 9789283224310. [Google Scholar]
- Schmitz, R.; Stanelle, J.; Hansmann, M.-L.; Küppers, R. Pathogenesis of Classical and Lymphocyte-Predominant Hodgkin Lymphoma. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 151–174. [Google Scholar] [CrossRef] [PubMed]
- Ushmorov, A.; Leithäuser, F.; Sakk, O.; Weinhaüsel, A.; Popov, S.W.; Möller, P.; Wirth, T. Epigenetic processes play a major role in B-cell-specific gene silencing in classical Hodgkin lymphoma. Blood 2006, 107, 2493–2500. [Google Scholar] [CrossRef] [PubMed]
- Thévenin, C.; Lucas, B.P.; Kozlow, E.J.; Kehrl, J.H. Cell type- and stage-specific expression of the CD20/B1 antigen correlates with the activity of a diverged octamer DNA motif present in its promote. J. Biol. Chem. 1993, 268, 5949–5956. [Google Scholar] [CrossRef]
- Young, R.L.; Korsmeyer, S.J. A Negative Regulatory Element in the bcl-2 5′-Untranslated Region Inhibits Expression from an Upstream Promoter. Mol. Cell. Biol. 1993, 13, 3686–3697. [Google Scholar] [CrossRef]
- Kozmik, Z.; Wang, S.; Dörfler, P.; Adams, B.; Busslinger, M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol. 1992, 12, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- DeKoter, R.P.; Singh, H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000, 288, 1439–1441. [Google Scholar] [CrossRef]
- Stein, H.; Marafioti, T.; Foss, H.D.; Laumen, H.; Hummel, M.; Anagnostopoulos, I.; Wirth, T.; Demel, G.; Falini, B. Down-regulation of BOB.1/OBF.1 and Oct2 in classical Hodgkin disease but not in lymphocyte predominant Hodgkin disease correlates with immunoglobulin transcription. Blood 2001, 97, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Mathas, S.; Janz, M.; Hummel, F.; Hummel, M.; Wollert-Wulf, B.; Lusatis, S.; Anagnostopoulos, I.; Lietz, A.; Sigvardsson, M.; Jundt, F.; et al. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat. Immunol. 2006, 7, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Jundt, F.; Anagnostopoulos, I.; Förster, R.; Mathas, S.; Stein, H.; Dörken, B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood 2002, 99, 3398–3403. [Google Scholar] [CrossRef] [PubMed]
- Foss, H.-D.; Reusch, R.; Demel, G.; Lenz, G.; Anagnostopoulos, I.; Hummel, M.; Stein, H. Frequent Expression of the B-Cell–Specific Activator Protein in Reed-Sternberg Cells of Classical Hodgkin’s Disease Provides Further Evidence for Its B-Cell Origin. Blood 1999, 94, 3108–3113. [Google Scholar] [CrossRef]
- Weniger, M.A.; Küppers, R. NF-κB deregulation in Hodgkin lymphoma. Semin. Cancer Biol. 2016, 39, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Lorenzon, D.; Cattaruzza, L.; Pinto, A.; Gloghini, A.; Carbone, A.; Colombatti, A. Expression of CCR5 receptors on Reed-Sternberg cells and Hodgkin lymphoma cell lines: Involvement of CCL5/Rantes in tumor cell growth and microenvironmental interactions. Int. J. Cancer 2008, 122, 769–776. [Google Scholar] [CrossRef]
- Chiu, A.; Xu, W.; He, B.; Dillon, S.R.; Gross, J.A.; Sievers, E.; Qiao, X.; Santini, P.; Hyjek, E.; Lee, J.W.; et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood 2007, 109, 729–739. [Google Scholar] [CrossRef]
- Fiumara, P.; Snell, V.; Li, Y.; Mukhopadhyay, A.; Younes, M.; Gillenwater, A.M.; Cabanillas, F.; Aggarwal, B.B.; Younes, A. Functional expression of receptor activator of nuclear factor kappaB in Hodgkin disease cell lines. Blood 2001, 98, 2784–2790. [Google Scholar] [CrossRef] [PubMed]
- Schwab, U.; Stein, H.; Gerdes, J.; Lemke, H.; Kirchner, H.; Schaadt, M.; Diehl, V. Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin’s disease and a subset of normal lymphoid cells. Nature 1982, 299, 65–67. [Google Scholar] [CrossRef]
- Cabannes, E.; Khan, G.; Aillet, F.; Jarrett, R.F.; Hay, R.T. Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IκBα. Oncogene 1999, 18, 3063–3070. [Google Scholar] [CrossRef]
- Emmerich, F.; Theurich, S.; Hummel, M.; Haeffker, A.; Vry, M.S.; Döhner, K.; Bommert, K.; Stein, H.; Dörken, B. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J. Pathol. 2003, 201, 413–420. [Google Scholar] [CrossRef]
- Jungnickel, B.; Staratschek-Jox, A.; Bräuninger, A.; Spieker, T.; Wolf, J.; Diehl, V.; Hansmann, M.L.; Rajewsky, K.; Küppers, R. Clonal deleterious mutations in the iκbα gene in the malignant cells in Hodgkin’s lymphoma. J. Exp. Med. 2000, 191, 395–401. [Google Scholar] [CrossRef]
- Otto, C.; Giefing, M.; Massow, A.; Vater, I.; Gesk, S.; Schlesner, M.; Richter, J.; Klapper, W.; Hansmann, M.-L.; Siebert, R.; et al. Genetic lesions of the TRAF3 and MAP3K14 genes in classical Hodgkin lymphoma. Br. J. Haematol. 2012, 157, 702–708. [Google Scholar] [CrossRef]
- Martín-Subero, J.I.; Gesk, S.; Harder, L.; Sonoki, T.; Tucker, P.W.; Schlegelberger, B.; Grote, W.; Novo, F.J.; Calasanz, M.J.; Hansmann, M.L.; et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 2002, 99, 1474–1477. [Google Scholar] [CrossRef]
- Scheeren, F.A.; Diehl, S.A.; Smit, L.A.; Beaumont, T.; Naspetti, M.; Bende, R.J.; Blom, B.; Karube, K.; Ohshima, K.; van Noesel, C.J.M.; et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: Evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood 2008, 111, 4706–4715. [Google Scholar] [CrossRef] [PubMed]
- Baus, D.; Pfitzner, E. Specific function of STAT3, SOCS1, and SOCS3 in the regulation of proliferation and survival of classical Hodgkin lymphoma cells. Int. J. Cancer 2006, 118, 1404–1413. [Google Scholar] [CrossRef]
- Kube, D.; Holtick, U.; Vockerodt, M.; Ahmadi, T.; Haier, B.; Behrmann, I.; Heinrich, P.C.; Diehl, V.; Tesch, H. STAT3 is constitutively activated in Hodgkin cell lines. Blood 2001, 98, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F.; Elia, A.J.; Gascoyne, R.D.; Patterson, B.; Trumper, L.; Kapp, U.; Mak, T.W. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002, 99, 618–626. [Google Scholar] [CrossRef]
- Dutton, A.; Reynolds, G.M.; Dawson, C.W.; Young, L.S.; Murray, P.G. Constitutive activation of phosphatidyl-inositide 3 kinase contributes to the survival of Hodgkin’s lymphoma cells through a mechanism involving Akt kinase and mTOR. J. Pathol. 2005, 205, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Georgakis, G.V.; Li, Y.; Rassidakis, G.Z.; Medeiros, L.J.; Mills, G.B.; Younes, A. Inhibition of the phosphatidylinositol-3 kinase/Akt promotes G1 cell cycle arrest and apoptosis in Hodgkin lymphoma. Br. J. Haematol. 2005, 132, 503–511. [Google Scholar] [CrossRef]
- Renne, C.; Willenbrock, K.; Küppers, R.; Hansmann, M.-L.; Bräuninger, A. Autocrine- and paracrine-activated receptor tyrosine kinases in classic Hodgkin lymphoma. Blood 2005, 105, 4051–4059. [Google Scholar] [CrossRef]
- Zheng, B.; Fiumara, P.; Li, Y.V.; Georgakis, G.; Snell, V.; Younes, M.; Vauthey, J.N.; Carbone, A.; Younes, A. MEK/ERK pathway is aberrantly active in Hodgkin disease: A signaling pathway shared by CD30, CD40, and RANK that regulates cell proliferation and survival. Blood 2003, 102, 1019–1027. [Google Scholar] [CrossRef]
- Aldinucci, D.; Celegato, M.; Casagrande, N. Microenvironmental interactions in classical Hodgkin lymphoma and their role in promoting tumor growth, immune escape and drug resistance. Cancer Lett. 2016, 380, 243–252. [Google Scholar] [CrossRef]
- Küppers, R. The biology of Hodgkin’s lymphoma. Nat. Rev. Cancer 2009, 9, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Gloghini, A.; Pinto, A.; De Filippi, R.; Carbone, A. The classical Hodgkin’s lymphoma microenvironment and its role in promoting tumour growth and immune escape. J. Pathol. 2010, 221, 248–263. [Google Scholar] [CrossRef]
- Samoszuk, M.; Nansen, L. Detection of interleukin-5 messenger RNA in Reed-Sternberg cells of Hodgkin’s disease with eosinophilia. Blood 1990, 75, 13–16. [Google Scholar] [CrossRef]
- Hanamoto, H.; Nakayama, T.; Miyazato, H.; Takegawa, S.; Hieshima, K.; Tatsumi, Y.; Kanamaru, A.; Yoshie, O. Expression of CCL28 by Reed-Sternberg cells defines a major subtype of classical Hodgkin’s disease with frequent infiltration of eosinophils and/or plasma cells. Am. J. Pathol. 2004, 164, 997–1006. [Google Scholar] [CrossRef]
- Cattaruzza, L.; Gloghini, A.; Olivo, K.; Di Francia, R.; Lorenzon, D.; De Filippi, R.; Carbone, A.; Colombatti, A.; Pinto, A.; Aldinucci, D. Functional coexpression of Interleukin (IL)-7 and its receptor (IL-7R) on Hodgkin and Reed-Sternberg cells: Involvement of IL-7 in tumor cell growth and microenvironmental interactions of Hodgkin’s lymphoma. Int. J. Cancer 2009, 125, 1092–1101. [Google Scholar] [CrossRef]
- Fischer, M.; Juremalm, M.; Olsson, N.; Backlin, C.; Sundström, C.; Nilsson, K.; Enblad, G.; Nilsson, G. Expression of CCL5/RANTES by Hodgkin and reed-sternberg cells and its possible role in the recruitment of mast cells into lymphomatous tissue. Int. J. Cancer 2003, 107, 197–201. [Google Scholar] [CrossRef]
- Baumforth, K.R.N.; Birgersdotter, A.; Reynolds, G.M.; Wei, W.; Kapatai, G.; Flavell, J.R.; Kalk, E.; Piper, K.; Lee, S.; Machado, L.; et al. Expression of the Epstein-Barr virus-encoded Epstein-Barr virus nuclear antigen 1 in Hodgkin’s lymphoma cells mediates Up-regulation of CCL20 and the migration of regulatory T cells. Am. J. Pathol. 2008, 173, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer 2005, 5, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Olivo, K.; Lorenzon, D.; Poletto, D.; Gloghini, A.; Carbone, A.; Pinto, A. The role of interleukin-3 in classical Hodgkin’s disease. Leuk. Lymphoma 2005, 46, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef] [PubMed]
- Theill, L.E.; Boyle, W.J.; Penninger, J.M. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu. Rev. Immunol. 2002, 20, 795–823. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.M.; Hsu, P.L. The nature of Reed-Sternberg cells: Phenotype, genotype, and other properties. Crit. Rev. Oncog. 1994, 5, 213–245. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.; Qu, Z.; Cardillo, T.M.; Chen, S.; Rosario, A.; Horak, I.D.; Hansen, H.J.; Goldenberg, D.M. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood 2004, 104, 3705–3711. [Google Scholar] [CrossRef] [PubMed]
- Foss, H.D.; Herbst, H.; Gottstein, S.; Demel, G.; Araujó, I.; Stein, H. Interleukin-8 in Hodgkin’s disease. Preferential expression by reactive cells and association with neutrophil density. Am. J. Pathol. 1996, 148, 1229–1236. [Google Scholar]
- Luciani, M.G.; Stoppacciaro, A.; Peri, G.; Mantovani, A.; Ruco, L.P. The monocyte chemotactic protein a (MCP-1) and interleukin 8 (IL-8) in Hodgkin’s disease and in solid tumours. Mol. Pathol. 1998, 51, 273–276. [Google Scholar] [CrossRef]
- Enblad, G.; Molin, D.; Glimelius, I.; Fischer, M.; Nilsson, G. The Potential Role of Innate Immunity in the Pathogenesis of Hodgkin’s Lymphoma. Hematol. Oncol. Clin. N. Am. 2007, 21, 805–823. [Google Scholar] [CrossRef]
- Wein, F.; Küppers, R. The role of T cells in the microenvironment of Hodgkin lymphoma. J. Leukoc. Biol. 2015, 99, 1–6. [Google Scholar] [CrossRef]
- Liu, Y.; Sattarzadeh, A.; Diepstra, A.; Visser, L.; van den Berg, A. The microenvironment in classical Hodgkin lymphoma: An actively shaped and essential tumor component. Semin. Cancer Biol. 2014, 24, 15–22. [Google Scholar] [CrossRef]
- Aldinucci, D.; Gloghini, A.; Pinto, A.; Colombatti, A.; Carbone, A. The role of CD40/CD40L and interferon regulatory factor 4 in Hodgkin lymphoma microenvironment. Leuk. Lymphoma 2012, 53, 195–201. [Google Scholar] [CrossRef]
- Veldman, J.; Visser, L.; Huberts-Kregel, M.; Muller, N.; Hepkema, B.; van den Berg, A.; Diepstra, A. Rosetting T cells in Hodgkin lymphoma are activated by immunological synapse components HLA class II and CD58. Blood 2020, 136, 2437–2441. [Google Scholar] [CrossRef]
- Poppema, S.; Potters, M.; Emmens, R.; Visser, L.; van den Berg, A. Immune reactions in classical Hodgkin’s lymphoma. Semin. Hematol. 1999, 36, 253–259. [Google Scholar]
- Skinnider, B.F.; Mak, T.W. The Role of Cytokines in Classical Hodgkin Lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Owen, A.; Iqbal, S.; Lee, A.; Matthews, J.; Wilson, A.; Calaminici, M.; Gribben, J.G. Defining characteristics of classical Hodgkin lymphoma microenvironment T-helper cells. Blood 2013, 122, 2856–2863. [Google Scholar] [CrossRef] [PubMed]
- Álvaro, T.; Lejeune, M.; Salvadó, M.T.; Bosch, R.; García, J.F.; Jaén, J.; Banham, A.H.; Roncador, G.; Montalbán, C.; Piris, M.A. Outcome in Hodgkin’s lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin. Cancer Res. 2005, 11, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Poppema, S.; Bhan, A.K.; Reinherz, E.L.; Posner, M.R.; Schlossman, S.F. In situ immunologic characterization of cellular constituents in lymph nodes and spleens involved by Hodgkin’s disease. Blood 1982, 59, 226–232. [Google Scholar] [CrossRef]
- Alvaro-Naranjo, T.; Lejeune, M.; Salvadó-Usach, M.T.; Bosch-Príncep, R.; Reverter-Branchat, G.; Jaén-Martínez, J.; Pons-Ferré, L.E. Tumor-infiltrating cells as a prognostic factor in Hodgkin’s lymphoma: A quantitative tissue microarray study in a large retrospective cohort of 267 patients. Leuk. Lymphoma 2005, 46, 1581–1591. [Google Scholar] [CrossRef]
- Kelley, T.W.; Pohlman, B.; Elson, P.; Hsi, E.D. The ratio of FOXP3+ regulatory T cells to granzyme B+ cytotoxic T/NK cells predicts prognosis in classical Hodgkin lymphoma and is independent of bcl-2 and MAL expression. Am. J. Clin. Pathol. 2007, 128, 958–965. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Cretney, E.; Kallies, A.; Nutt, S.L. Differentiation and function of Foxp3+ effector regulatory T cells. Trends Immunol. 2013, 34, 74–80. [Google Scholar] [CrossRef]
- Shevach, E.M. Mechanisms of Foxp3+ T Regulatory Cell-Mediated Suppression. Immunity 2009, 30, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Bluestone, J.A. The Foxp3+ regulatory T cell: A jack of all trades, master of regulation. Nat. Immunol. 2008, 9, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A.A.; Collison, L.W.; Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008, 8, 523–532. [Google Scholar] [CrossRef]
- Bates, G.J.; Fox, S.B.; Han, C.; Leek, R.D.; Garcia, J.F.; Harris, A.L.; Banham, A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 2006, 24, 5373–5380. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Gao, Q.; Qiu, S.-J.; Fan, J.; Zhou, J.; Wang, X.-Y.; Xiao, Y.-S.; Xu, Y.; Li, Y.-W.; Tang, Z.-Y. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J. Clin. Oncol. 2007, 25, 2586–2593. [Google Scholar] [CrossRef]
- Petersen, R.P.; Campa, M.J.; Sperlazza, J.; Conlon, D.; Joshi, M.-B.; Harpole, D.H.; Patz, E.F. Tumor infiltrating Foxp3+ regulatory T-cells are associated with recurrence in pathologic stage I NSCLC patients. Cancer 2006, 107, 2866–2872. [Google Scholar] [CrossRef]
- Perrone, G.; Ruffini, P.A.; Catalano, V.; Spino, C.; Santini, D.; Muretto, P.; Spoto, C.; Zingaretti, C.; Sisti, V.; Alessandroni, P.; et al. Intratumoural FOXP3-positive regulatory T cells are associated with adverse prognosis in radically resected gastric cancer. Eur. J. Cancer 2008, 44, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Shah, W.; Yan, X.; Jing, L.; Zhou, Y.; Chen, H.; Wang, Y. A reversed CD4/CD8 ratio of tumor-infiltrating lymphocytes and a high percentage of CD4+FOXP3+ regulatory T cells are significantly associated with clinical outcome in squamous cell carcinoma of the cervix. Cell. Mol. Immunol. 2011, 8, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Vardhana, S.; Younes, A. The immune microenvironment in hodgkin lymphoma: T cells, B cells, and immune checkpoints. Haematologica 2016, 101, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Chetaille, B.; Bertucci, F.; Finetti, P.; Esterni, B.; Stamatoullas, A.; Picquenot, J.M.; Copin, M.C.; Morschhauser, F.; Casasnovas, O.; Petrella, T.; et al. Molecular profiling of classical Hodgkin lymphoma tissues uncovers variations in the tumor microenvironment and correlations with EBV infection and outcome. Blood 2009, 113, 2765–2775. [Google Scholar] [CrossRef]
- Inoue, S.; Leitner, W.W.; Golding, B.; Scott, D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006, 66, 7741–7747. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-Associated Macrophages and Survival in Classic Hodgkin’s Lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Azambuja, D.; Natkunam, Y.; Biasoli, I.; Lossos, I.S.; Anderson, M.W.; Morais, J.C.; Spector, N. Lack of association of tumor-associated macrophages with clinical outcome in patients with classical Hodgkin’s lymphoma. Ann. Oncol. 2012, 23, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.A.; Jain, S.; Ren, Q.; Zarineh, A.; Liu, C.; Ibrahim, S. CD163 versus CD68 in tumor associated macrophages of classical hodgkin lymphoma. Diagn. Pathol. 2012, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Kayal, S.; Mathur, S.; Karak, A.K.; Kumar, L.; Sharma, A.; Bakhshi, S.; Raina, V. CD68 tumor-associated macrophage marker is not prognostic of clinical outcome in classical Hodgkin lymphoma. Leuk. Lymphoma 2014, 55, 1031–1037. [Google Scholar] [CrossRef]
- Tan, K.L.; Scott, D.W.; Hong, F.; Kahl, B.S.; Fisher, R.I.; Bartlett, N.L.; Advani, R.H.; Buckstein, R.; Rimsza, L.M.; Connors, J.M.; et al. Tumor-associated macrophages predict inferior outcomes in classic Hodgkin lymphoma: A correlative study from the E2496 Intergroup trial. Blood 2012, 120, 3280–3287. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Coutinho, R.; Wilson, A.; Matthews, J.; Owen, A.; Shanyinde, M.; Lister, T.A.; Calaminici, M.; Gribben, J.G. Expression of FOXP3, CD68, and CD20 at Diagnosis in the Microenvironment of Classical Hodgkin Lymphoma Is Predictive of Outcome. J. Clin. Oncol. 2013, 31, 256–262. [Google Scholar] [CrossRef]
- Tzankov, A.; Matter, M.S.; Dirnhofer, S. Refined Prognostic Role of CD68-Positive Tumor Macrophages in the Context of the Cellular Micromilieu of Classical Hodgkin Lymphoma. Pathobiology 2010, 77, 301–308. [Google Scholar] [CrossRef]
- Gotti, M.; Nicola, M.; Lucioni, M.; Fiaccadori, V.; Ferretti, V.; Sciarra, R.; Costanza, M.; Bono, E.; Molo, S.; Maffi, A.; et al. Independent prognostic impact of tumour-infiltrating macrophages in early-stage Hodgkin’s lymphoma. Hematol. Oncol. 2016, 35, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Diepstra, A.; Lee, T.; Chan, F.C.; Farinha, P.; Tan, K.; Telenius, A.; Barclay, L.; Shah, S.P.; Connors, J.M.; et al. Gene expression profiling of microdissected Hodgkin Reed-Sternberg cells correlates with treatment outcome in classical Hodgkin lymphoma. Blood 2012, 120, 3530–3540. [Google Scholar] [CrossRef] [PubMed]
- Allavena, P.; Mantovani, A. Immunology in the clinic review series; focus on cancer: Tumour-associated macrophages: Undisputed stars of the inflammatory tumour microenvironment. Clin. Exp. Immunol. 2012, 167, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.O.; Schmidt, H.; Møller, H.J.; Høyer, M.; Maniecki, M.B.; Sjoegren, P.; Christensen, I.J.; Steiniche, T. Macrophage markers in serum and tumor have prognostic impact in American Joint Committee on Cancer stage I/II melanoma. J. Clin. Oncol. 2009, 27, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, L.; Che, G.; Yu, N.; Dai, F.; You, Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer 2010, 10, 112. [Google Scholar] [CrossRef]
- Zaki, M.A.A.; Wada, N.; Ikeda, J.; Shibayama, H.; Hashimoto, K.; Yamagami, T.; Tatsumi, Y.; Tsukaguchi, M.; Take, H.; Tsudo, M.; et al. Prognostic implication of types of tumor-associated macrophages in Hodgkin lymphoma. Virchows Arch. 2011, 459, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef]
- Molin, D.; Edstrom, A.; Glimelius, I.; Glimelius, B.; Nilsson, G.; Sundstrom, C.; Enblad, G. Mast cell infiltration correlates with poor prognosis in Hodgkin’s lymphoma. Br. J. Haematol. 2002, 119, 122–124. [Google Scholar] [CrossRef]
- Molin, D.; Fischer, M.; Xiang, Z.; Larsson, U.; Harvima, I.; Venge, P.; Nilsson, K.; Sundström, C.; Enblad, G.; Nilsson, G. Mast cells express functional CD30 ligand and are the predominant CD30L-positive cells in Hodgkin’s disease. Br. J. Haematol. 2001, 114, 616–623. [Google Scholar] [CrossRef]
- Mizuno, H.; Nakayama, T.; Miyata, Y.; Saito, S.; Nishiwaki, S.; Nakao, N.; Takeshita, K.; Naoe, T. Mast cells promote the growth of Hodgkin’s lymphoma cell tumor by modifying the tumor microenvironment that can be perturbed by bortezomib. Leukemia 2012, 26, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Oldford, S.A.; Marshall, J.S. Mast cells as targets for immunotherapy of solid tumors. Mol. Immunol. 2015, 63, 113–124. [Google Scholar] [CrossRef]
- Keresztes, K.; Szollosi, Z.; Simon, Z.; Tarkanyi, I.; Nemes, Z.; Illes, A. Retrospective Analysis of the Prognostic Role of Tissue Eosinophil and Mast Cells in Hodgkin’s Lymphoma. Pathol. Oncol. Res. 2007, 13, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.D.; Kamper, P.; Nielsen, P.S.; Bendix, K.; Riber-Hansen, R.; Steiniche, T.; Hamilton-Dutoit, S.; Clausen, M.; d’Amore, F. Tumour-associated mast cells in classical Hodgkin’s lymphoma: Correlation with histological subtype, other tumour-infiltrating inflammatory cell subsets and outcome. Eur. J. Haematol. 2016, 96, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Poppema, S. Immunobiology and pathophysiology of Hodgkin lymphomas. Hematol. Am. Soc. Hematol. Educ. Progr. 2005, 2005, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Diepstra, A.; Poppema, S.; Boot, M.; Visser, L.; Nolte, I.M.; Niens, M.; Te Meerman, G.J.; Van Den Berg, A. HLA-G protein expression as a potential immune escape mechanism in classical Hodgkin’s lymphoma. Tissue Antigens 2008, 71, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Altomonte, M.; Gloghini, A.; Bertola, G.; Gasparollo, A.; Carbone, A.; Ferrone, S.; Maio, M. Differential expression of cell adhesion molecules CD54/CD11a and CD58/CD2 by human melanoma cells and functional role in their interaction with cytotoxic cells. Cancer Res. 1993, 53, 3343–3348. [Google Scholar] [PubMed]
- Gwin, J.L.; Gercel-Taylor, C.; Taylor, D.D.; Eisenberg, B. Role of LFA-3, ICAM-1, and MHC Class I on the Sensitivity of Human Tumor Cells to LAK Cells. J. Surg. Res. 1996, 60, 129–136. [Google Scholar] [CrossRef]
- Schneider, M.; Schneider, S.; Zühlke-Jenisch, R.; Klapper, W.; Sundström, C.; Hartmann, S.; Hansmann, M.-L.; Siebert, R.; Küppers, R.; Giefing, M. Alterations of the CD58 gene in classical Hodgkin lymphoma. Genes. Chromosomes Cancer 2015, 54, 638–645. [Google Scholar] [CrossRef]
- Reichel, J.; Chadburn, A.; Rubinstein, P.G.; Giulino-Roth, L.; Tam, W.; Liu, Y.; Gaiolla, R.; Eng, K.; Brody, J.; Inghirami, G.; et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015, 125, 1061–1072. [Google Scholar] [CrossRef]
- Liu, Y.; Abdul Razak, F.R.; Terpstra, M.; Chan, F.C.; Saber, A.; Nijland, M.; van Imhoff, G.; Visser, L.; Gascoyne, R.; Steidl, C.; et al. The mutational landscape of Hodgkin lymphoma cell lines determined by whole-exome sequencing. Leukemia 2014, 28, 2248–2251. [Google Scholar] [CrossRef]
- Abdul Razak, F.R.; Diepstra, A.; Visser, L.; van den Berg, A. CD58 mutations are common in Hodgkin lymphoma cell lines and loss of CD58 expression in tumor cells occurs in Hodgkin lymphoma patients who relapse. Genes Immun. 2016, 17, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.T.; Pang, W.L.; Chong, S.M.; Castella, A.; Al-Salam, S.; Tan, T.E.; Moh, M.C.; Koh, L.K.; Gan, S.U.; Cheng, C.K.; et al. Expression of CD137 on Hodgkin and Reed-Sternberg cells inhibits T-cell activation by eliminating CD137 ligand expression. Cancer Res. 2013, 73, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Chemnitz, J.M.; Driesen, J.; Classen, S.; Riley, J.L.; Debey, S.; Beyer, M.; Popov, A.; Zander, T.; Schultze, J.L. Prostaglandin E2 Impairs CD4+ T Cell Activation by Inhibition of lck: Implications in Hodgkin’s Lymphoma. Cancer Res. 2006, 66, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Juszczynski, P.; Ouyang, J.; Monti, S.; Rodig, S.J.; Takeyama, K.; Abramson, J.; Chen, W.; Kutok, J.L.; Rabinovich, G.A.; Shipp, M.A. The AP1-dependent secretion of galectin-1 by Reed Sternberg cells fosters immune privilege in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA 2007, 104, 13134–13139. [Google Scholar] [CrossRef] [PubMed]
- Spear, P.; Wu, M.-R.; Sentman, M.-L.; Sentman, C.L. NKG2D ligands as therapeutic targets. Cancer Immun. 2013, 13, 8. [Google Scholar]
- Maggio, E.M.; Van Den Berg, A.; de Jong, D.; Diepstra, A.; Poppema, S. Low frequency of FAS mutations in Reed-Sternberg cells of Hodgkin’s lymphoma. Am. J. Pathol. 2003, 162, 29–35. [Google Scholar] [CrossRef]
- Thomas, R.K.; Kallenborn, A.; Wickenhauser, C.; Schultze, J.L.; Draube, A.; Vockerodt, M.; Re, D.; Diehl, V.; Wolf, J. Constitutive Expression of c-FLIP in Hodgkin and Reed-Sternberg Cells. Am. J. Pathol. 2002, 160, 1521–1528. [Google Scholar] [CrossRef]
- Mathas, S.; Lietz, A.; Anagnostopoulos, I.; Hummel, F.; Wiesner, B.; Janz, M.; Jundt, F.; Hirsch, B.; Jöhrens-Leder, K.; Vornlocher, H.-P.; et al. c-FLIP mediates resistance of Hodgkin/Reed-Sternberg cells to death receptor-induced apoptosis. J. Exp. Med. 2004, 199, 1041–1052. [Google Scholar] [CrossRef]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef]
- Yamamoto, R.; Nishikori, M.; Kitawaki, T.; Sakai, T.; Hishizawa, M.; Tashima, M.; Kondo, T.; Ohmori, K.; Kurata, M.; Hayashi, T.; et al. PD-1 PD-1 ligand interaction contributes to immunosuppressive microenvironment of Hodgkin lymphoma. Blood 2008, 111, 3220–3224. [Google Scholar] [CrossRef] [PubMed]
- Roemer, M.G.M.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J. Clin. Oncol. 2016, 19, 3462–3473. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 activity and EBV infection induce PD-l1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011, 471, 377–381. [Google Scholar] [CrossRef]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Śledzińska, A.; Menger, L.; Bergerhoff, K.; Peggs, K.S.; Quezada, S.A. Negative immune checkpoints on T lymphocytes and their relevance to cancer immunotherapy. Mol. Oncol. 2015, 9, 1936–1965. [Google Scholar] [CrossRef] [PubMed]
- Devilard, E.; Bertucci, F.; Trempat, P.; Bouabdallah, R.; Loriod, B.; Giaconia, A.; Brousset, P.; Granjeaud, S.; Nguyen, C.; Birnbaum, D.; et al. Gene expression profiling defines molecular subtypes of classical Hodgkin’s disease. Oncogene 2002, 21, 3095–3102. [Google Scholar] [CrossRef]
- Sanchez-Aguilera, A.; Montalbán, C.; de la Cueva, P.; Sánchez-Verde, L.; Morente, M.M.; García-Cosío, M.; García-Laraña, J.; Bellas, C.; Provencio, M.; Romagosa, V.; et al. Tumor microenvironment and mitotic checkpoint are key factors in the outcome of classic Hodgkin lymphoma. Blood 2006, 108, 662–668. [Google Scholar] [CrossRef]
- Scott, D.W.; Chan, C.; Hong, F.; Rogic, S.; Tan, K.L.; Meissner, B.; Ben-Neriah, S.; Boyle, M.; Kridel, R.; Telenius, A.; et al. Gene Expression-Based Model Using Formalin-Fixed Paraffin-Embedded Biopsies Predicts Overall Survival in Advanced-Stage Classical Hodgkin Lymphoma. J. Clin. Oncol. 2012, 31, 692–700. [Google Scholar] [CrossRef]
- Jachimowicz, R.D.; Klapper, W.; Glehr, G.; Müller, H.; Haverkamp, H.; Thorns, C.; Hansmann, M.L.; Möller, P.; Stein, H.; Rehberg, T.; et al. Gene expression-based outcome prediction in advanced stage classical Hodgkin lymphoma treated with BEACOPP. Leukemia 2021. [Google Scholar] [CrossRef]
- Chan, F.C.; Mottok, A.; Gerrie, A.S.; Power, M.; Savage, K.J.; Shah, S.P.; Connors, J.M.; Gascoyne, R.D.; Scott, D.W.; Steidl, C.; et al. Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical hodgkin lymphoma. J. Clin. Oncol. 2017, 35, 3722–3733. [Google Scholar] [CrossRef] [PubMed]
- Calvente, L.; Tremblay-LeMay, R.; Xu, W.; Chan, F.C.; Hong, M.; Zhang, T.; Yhim, H.Y.; Kuruvilla, J.; Crump, M.; Kukreti, V.; et al. Validation of the RHL30 digital gene expression assay as a prognostic biomarker for relapsed Hodgkin lymphoma. Br. J. Haematol. 2020, 190, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Luminari, S.; Donati, B.; Casali, M.; Valli, R.; Santi, R.; Puccini, B.; Kovalchuk, S.; Ruffini, A.; Fama, A.; Berti, V.; et al. A Gene Expression–based Model to Predict Metabolic Response after Two Courses of ABVD in Hodgkin Lymphoma Patients. Clin. Cancer Res. 2020, 26, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Donati, B.; Ferrari, A.; Ruffini, A.; Manzotti, G.; Fragliasso, V.; Merli, F.; Zanelli, M.; Valli, R.; Luminari, S.; Ciarrocchi, A. Gene expression profile unveils diverse biological effect of serum vitamin D in Hodgkin’s and diffuse large B-cell lymphoma. Hematol. Oncol. 2021, 39, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Diehl, V.; Franklin, J.; Pfreundschuh, M.; Lathan, B.; Paulus, U.; Hasenclever, D.; Tesch, H.; Herrmann, R.; Dörken, B.; Müller-Hermelink, H.-K.; et al. Standard and increased-dose BEACOPP chemotherapy compared with COPP-ABVD for advanced Hodgkin’s disease. N. Engl. J. Med. 2003, 348, 2386–2395. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.; McKenzie, H. How I treat advanced classical Hodgkin lymphoma. Blood 2015, 125, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Duggan, D.B.; Petroni, G.R.; Johnson, J.L.; Glick, J.H.; Fisher, R.I.; Connors, J.M.; Canellos, G.P.; Peterson, B.A. Randomized comparison of ABVD and MOPP/ABV hybrid for the treatment of advanced Hodgkin’s disease: Report of an intergroup trial. J. Clin. Oncol. 2003, 21, 607–614. [Google Scholar] [CrossRef]
- Schmitz, N.; Pfistner, B.; Sextro, M.; Sieber, M.; Carella, A.M.; Haenel, M.; Boissevain, F.; Zschaber, R.; Müller, P.; Kirchner, H.; et al. Aggressive conventional chemotherapy compared with high-dose chemotherapy with autologous haemopoietic stem-cell transplantation for relapsed chemosensitive Hodgkin’s disease: A randomised trial. Lancet 2002, 359, 2065–2071. [Google Scholar] [CrossRef]
- Younes, A.; Bartlett, N.L.; Leonard, J.P.; Kennedy, D.A.; Lynch, C.M.; Sievers, E.L.; Forero-Torres, A. Brentuximab Vedotin (SGN-35) for Relapsed CD30-Positive Lymphomas. N. Engl. J. Med. 2010, 363, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, A.J.; Schöder, H.; Yahalom, J.; McCall, S.J.; Fox, S.Y.; Gerecitano, J.; Grewal, R.; Hamlin, P.A.; Horwitz, S.; Kobos, R.; et al. PET-adapted sequential salvage therapy with brentuximab vedotin followed by augmented ifosamide, carboplatin, and etoposide for patients with relapsed and refractory Hodgkin’s lymphoma: A non-randomised, open-label, single-centre, phase 2 study. Lancet Oncol. 2015, 16, 284–292. [Google Scholar] [CrossRef]
- Kumar, A.; Casulo, C.; Yahalom, J.; Schöder, H.; Barr, P.M.; Caron, P.; Chiu, A.; Constine, L.S.; Drullinsky, P.; Friedberg, J.W.; et al. Brentuximab vedotin and AVD followed by involved-site radiotherapy in early stage, unfavorable risk Hodgkin lymphoma. Blood 2016, 128, 1458–1464. [Google Scholar] [CrossRef]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 Blockade with Nivolumab in Relapsed or Refractory Hodgkin’s Lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Younes, A.; Santoro, A.; Shipp, M.; Zinzani, P.L.; Timmerman, J.M.; Ansell, S.; Armand, P.; Fanale, M.; Ratanatharathorn, V.; Kuruvilla, J.; et al. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: A multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016, 17, 1283–1294. [Google Scholar] [CrossRef]
- Armand, P.; Shipp, M.A.; Ribrag, V.; Michot, J.M.; Zinzani, P.L.; Kuruvilla, J.; Snyder, E.S.; Ricart, A.D.; Balakumaran, A.; Rose, S.; et al. Programmed death-1 blockade with pembrolizumab in patients with classical hodgkin lymphoma after brentuximab vedotin failure. J. Clin. Oncol. 2016, 34, 3733–3739. [Google Scholar] [CrossRef]
- Moskowitz, C.H.; Zinzani, P.L.; Fanale, M.A.; Armand, P.; Johnson, N.A.; Radford, J.A.; Ribrag, V.; Molin, D.; Vassilakopoulos, T.P.; Tomita, A.; et al. Pembrolizumab in Relapsed/Refractory Classical Hodgkin Lymphoma: Primary End Point Analysis of the Phase 2 Keynote-087 Study. Blood 2016, 128, 1107. [Google Scholar] [CrossRef]
- Chen, R.; Zinzani, P.L.; Fanale, M.A.; Armand, P.; Johnson, N.A.; Brice, P.; Radford, J.; Ribrag, V.; Molin, D.; Vassilakopoulos, T.P.; et al. Phase II Study of the Efficacy and Safety of Pembrolizumab for Relapsed/Refractory Classic Hodgkin Lymphoma. J. Clin. Oncol. 2017, 35, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Armand, P.; Kuruvilla, J.; Michot, J.-M.; Ribrag, V.; Zinzani, P.L.; Zhu, Y.; Marinello, P.; Nahar, A.; Moskowitz, C.H. KEYNOTE-013 4-year follow-up of pembrolizumab in classical Hodgkin lymphoma after brentuximab vedotin failure. Blood Adv. 2020, 4, 2617–2622. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zinzani, P.L.; Lee, H.J.; Armand, P.; Johnson, N.A.; Brice, P.; Radford, J.; Ribrag, V.; Molin, D.; Vassilakopoulos, T.P.; et al. Pembrolizumab in Relapsed or Refractory Hodgkin Lymphoma: 2-Year Follow-Up of KEYNOTE-087. Blood 2019, 134, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Callahan, M.K.; Wolchok, J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015, 33, 1974–1982. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.L.; Leytze, G.M.; Emswiler, J.; Peach, R.; Bajorath, J.; Cosand, W.; Linsley, P.S. Covalent dimerization of CD28/CTLA-4 and oligomerization of CD80/CD86 regulate T cell costimulatory interactions. J. Biol. Chem. 1996, 271, 26762–26771. [Google Scholar] [CrossRef] [PubMed]
- Quandt, D.; Hoff, H.; Rudolph, M.; Fillatreau, S.; Brunner-Weinzierl, M.C. A new role of CTLA-4 on B cells in thymus-dependent immune responses in vivo. J. Immunol. 2007, 179, 7316–7324. [Google Scholar] [CrossRef]
- Bashey, A.; Medina, B.; Corringham, S.; Pasek, M.; Carrier, E.; Vrooman, L.; Lowy, I.; Solomon, S.R.; Morris, L.E.; Holland, H.K.; et al. CTLA4 blockade with ipilimumab to treat relapse of malignancy after allogeneic hematopoietic cell transplantation. Blood 2009, 113, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, C.S.; Connors, J.M.; Friedberg, J.W.; Leonard, J.P.; Kahl, B.S.; Little, R.F.; Baizer, L.; Evens, A.M.; Hoppe, R.T.; Kelly, K.M.; et al. Hodgkin lymphoma: Current status and clinical trial recommendations. J. Natl. Cancer Inst. 2017, 109, djw249. [Google Scholar] [CrossRef][Green Version]
- Mottok, A.; Steidl, C. Biology of classical Hodgkin lymphoma: Implications for prognosis and novel therapies. Blood 2018, 131, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef]
- Von Tresckow, B.; Morschhauser, F.; Ribrag, V.; Topp, M.S.; Chien, C.; Seetharam, S.; Aquino, R.; Kotoulek, S.; De Boer, C.J.; Engert, A. An Open-Label, Multicenter, Phase I/II Study of JNJ-40346527, a CSF-1R Inhibitor, in Patients with Relapsed or Refractory Hodgkin Lymphoma. Clin. Cancer Res. 2015, 21, 1843–1850. [Google Scholar] [CrossRef]
- Hombach, A.; Jung, W.; Pohl, C.; Renner, C.; Sahin, U.; Schmits, R.; Wolf, J.; Kapp, U.; Diehl, V.; Pfreundschuh, M. A CD16/CD30 bispecific monoclonal antibody induces lysis of hodgkin’s cells by unstimulated natural killer cells In AND In vivo. Int. J. Cancer 1993, 55, 830–836. [Google Scholar] [CrossRef]
- Rothe, A.; Sasse, S.; Topp, M.S.; Eichenauer, D.A.; Hummel, H.; Reiners, K.S.; Dietlein, M.; Kuhnert, G.; Kessler, J.; Buerkle, C.; et al. A phase 1 study of the bispecific anti-CD30/CD16A antibody construct AFM13 in patients with relapsed or refractory Hodgkin lymphoma. Blood 2015, 125, 4024–4031. [Google Scholar] [CrossRef]
- Ramos, C.A.; Ballard, B.; Zhang, H.; Dakhova, O.; Gee, A.P.; Mei, Z.; Bilgi, M.; Wu, M.F.; Liu, H.; Grilley, B.; et al. Clinical and immunological responses after CD30-specific chimeric antigen receptor-redirected lymphocytes. J. Clin. Investig. 2017, 127, 3462–3471. [Google Scholar] [CrossRef]
- Ruella, M.; Klichinsky, M.; Kenderian, S.S.; Shestova, O.; Ziober, A.; Kraft, D.O.; Feldman, M.; Wasik, M.A.; June, C.H.; Gill, S. Overcoming the immunosuppressive tumor microenvironment of Hodgkin lymphoma using chimeric antigen receptor T cells. Cancer Discov. 2017, 7, 1154–1167. [Google Scholar] [CrossRef] [PubMed]
- Fromm, J.R. Flow cytometric analysis of CD123 is useful for immunophenotyping classical Hodgkin lymphoma. Cytom. Part B Clin. Cytom. 2011, 80B, 91–99. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opinto, G.; Agostinelli, C.; Ciavarella, S.; Guarini, A.; Maiorano, E.; Ingravallo, G. Hodgkin Lymphoma: A Special Microenvironment. J. Clin. Med. 2021, 10, 4665. https://doi.org/10.3390/jcm10204665
Opinto G, Agostinelli C, Ciavarella S, Guarini A, Maiorano E, Ingravallo G. Hodgkin Lymphoma: A Special Microenvironment. Journal of Clinical Medicine. 2021; 10(20):4665. https://doi.org/10.3390/jcm10204665
Chicago/Turabian StyleOpinto, Giuseppina, Claudio Agostinelli, Sabino Ciavarella, Attilio Guarini, Eugenio Maiorano, and Giuseppe Ingravallo. 2021. "Hodgkin Lymphoma: A Special Microenvironment" Journal of Clinical Medicine 10, no. 20: 4665. https://doi.org/10.3390/jcm10204665
APA StyleOpinto, G., Agostinelli, C., Ciavarella, S., Guarini, A., Maiorano, E., & Ingravallo, G. (2021). Hodgkin Lymphoma: A Special Microenvironment. Journal of Clinical Medicine, 10(20), 4665. https://doi.org/10.3390/jcm10204665







