Effect of Age on Innate and Adaptive Immunity in Hospitalized COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Peripheral Blood Mononuclear Cell (PBMC) Isolation
2.3. Quantification of SARS-CoV-2
2.4. Viral Sequence
2.5. Cell Surface Staining
2.6. Inflammatory Cytokines
2.7. Biochemistry Assay
2.8. Serological Responses
3. Quantification and Statistical Analysis
Statistical Analysis
4. Results
4.1. Baseline Characteristics of the COVID-19 Study Population
4.2. Age Is Associated with Increased SARS-CoV-2 Viral Load, Plasma Inflammation Markers and Delayed Clinical Recovery
4.3. Elderly Patients Exhibited Reduced Monocyte Activation and Function
4.4. Dendritic Cells Impairment in Patients above 65 Years
4.5. Blunted Natural Killer Cell Activation and Lower IL-2 Levels in Older Patients
4.6. Reduced T Cell Activation in Older COVID-19 Patients
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sheahan, T.P.; Frieman, M.B. The continued epidemic threat of SARS-CoV-2 and implications for the future of global public health. Curr. Opin. Virol. 2020, 40, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Poonam; Rathi, B. Coronavirus Disease COVID-19: A New Threat to Public Health. Curr. Top. Med. Chem. 2020, 20, 599–600. [Google Scholar] [CrossRef]
- Abu-Zidan, F.M.; Khan, G.; Sheek-Hussein, M.; Alsuwaidi, A.; Idris, K. Novel coronavirus pandemic: A global health threat. Turk. J. Emerg. Med. 2020, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Yaya, S.; Otu, A.; Labonté, R. Globalisation in the time of COVID-19: Repositioning Africa to meet the immediate and remote challenges. Glob. Health 2020, 16, 1–7. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Morawska, L.; Milton, D.K. It Is Time to Address Airborne Transmission of Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2020, 71, 2311–2313. [Google Scholar] [CrossRef]
- He, D.; Zhao, S.; Lin, Q.; Zhuang, Z.; Cao, P.; Wang, M.H.; Yang, L. The relative transmissibility of asymptomatic COVID-19 infections among close contacts. Int. J. Infect. Dis. 2020, 94, 145–147. [Google Scholar] [CrossRef]
- Chen, H.; Chen, Y.; Lian, Z.; Wen, L.; Sun, B.; Wang, P.; Li, X.; Liu, Q.; Yu, X.; Lu, Y.; et al. Correlation between the migration scale index and the number of new confirmed coronavirus disease 2019 cases in China. Epidemiol. Infect. 2020, 148, 1–11. [Google Scholar] [CrossRef]
- Tsai, P.-H.; Lai, W.-Y.; Lin, Y.-Y.; Luo, Y.-H.; Chen, H.-K.; Chen, Y.-M.; Lai, Y.-C.; Kuo, L.-C.; Chen, S.-D.; Chang, K.-J.; et al. Clinical manifestation and disease progression in COVID-19 infection. J. Chin. Med. Assoc. 2021, 84, 3–8. [Google Scholar] [CrossRef]
- Anka, A.U.; Tahir, M.I.; Abubakar, S.D.; Alsabbagh, M.; Zian, Z.; Hamedifar, H.; Sabzevari, A.; Azizi, G. Coronavirus disease 2019 (COVID-19): An overview of the immunopathology, serological diagnosis and management. Scand. J. Immunol. 2021, 93, e12998. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; for the Gansu Provincial Medical Treatment Expert Group of COVID-19; Bai, X.; Wang, J.; Yu, Q.; Liu, W.; Pu, J.; Wang, X.; Hu, J.; Xu, D.; et al. Clinical characteristics of coronavirus disease 2019 in Gansu province, China. Ann. Palliat. Med. 2020, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Huang, Y.; Guo, Y.; Yin, M.; Chen, X.; Xiao, L.; Deng, G. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int. J. Infect. Dis. 2020, 96, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, P.; Zhu, J.; Zhong, Z.; Li, H.; Pang, J.; Li, B.; Zhang, J. Association of elevated inflammatory markers and severe COVID-19: A meta-analysis. Medicine 2020, 99, e23315. [Google Scholar] [CrossRef] [PubMed]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; van de Veen, W.; Bruggen, M.C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Schijns, V.; Lavelle, E.C. Prevention and treatment of COVID-19 disease by controlled modulation of innate immunity. Eur. J. Immunol. 2020, 50, 932–938. [Google Scholar] [CrossRef]
- Lee, S.; Channappanavar, R.; Kanneganti, T.-D. Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Trends Immunol. 2020, 41, 1083–1099. [Google Scholar] [CrossRef]
- Sallenave, J.M.; Guillot, L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in COVID-19: Key Therapeutic Targets? Front. Immunol. 2020, 11, 1229. [Google Scholar] [CrossRef]
- Park, S.H. An Impaired Inflammatory and Innate Immune Response in COVID-19. Mol. Cells 2021, 44, 384–391. [Google Scholar] [CrossRef]
- Taefehshokr, N.; Taefehshokr, S.; Hemmat, N.; Heit, B. COVID-19: Perspectives on Innate Immune Evasion. Front. Immunol. 2020, 11, 580641. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Portela Sousa, C.; Brites, C. Immune response in SARS-CoV-2 infection: The role of interferons type I and type III. Braz. J. Infect. Dis. 2020, 24, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response during SARS-CoV-2 Infection: Lessons from the Past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; To, K.K.; Wong, Y.C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.Y.; Lau, T.T.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e5. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.A.; Cascino, K.; Ordonez, A.A.; Zhou, W.; Vaghasia, A.; Hamacher-Brady, A.; Brady, N.R.; Sun, I.H.; Wang, R.; Rosenberg, A.Z.; et al. Mitochondrial induced T cell apoptosis and aberrant myeloid metabolic programs define distinct immune cell subsets during acute and recovered SARS-CoV-2 infection. MedRxiv 2020, 2020.09.10.20186064. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Hsin-Hui, H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature helps predict COVID-19 severity and death. MedRxiv 2020, 2020.05.28.20115758. [Google Scholar] [CrossRef]
- Niccoli, T.; Partridge, L. Ageing as a Risk Factor for Disease. Curr. Biol. 2012, 22, R741–R752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooqui, T.; Farooqui, A.A. Aging: An important factor for the pathogenesis of neurodegenerative diseases. Mech. Ageing Dev. 2009, 130, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Morsiani, C.; Conte, M.; Santoro, A.; Grignolio, A.; Monti, D.; Capri, M.; Salvioli, S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Nasi, M.; DE Biasi, S.; Gibellini, L.; Bianchini, E.; Pecorini, S.; Bacca, V.; Guaraldi, G.; Mussini, C.; Pinti, M.; Cossarizza, A. Ageing and inflammation in patients with HIV infection. Clin. Exp. Immunol. 2017, 187, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Simon, K.; Simon, R.; Serafińska, S. HIV/AIDS and aging. Prz. Epidemiol. 2010, 64, 287–292. [Google Scholar]
- Nikolich-Zugich, J. The twilight of immunity: Emerging concepts in aging of the immune system. Nat. Immunol. 2018, 19, 10–19. [Google Scholar] [CrossRef]
- Montecino-Rodriguez, E.; Berent-Maoz, B.; Dorshkind, K. Causes, consequences, and reversal of immune system aging. J. Clin. Investig. 2013, 123, 958–965. [Google Scholar] [CrossRef]
- Weyand, C.M.; Goronzy, J.J. Aging of the Immune System. Mechanisms and Therapeutic Targets. Ann. Am. Thorac. Soc. 2016, 13, S422–S428. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Klein, S.L.; Garibaldi, B.T.; Li, H.; Wu, C.; Osevala, N.M.; Li, T.; Margolick, J.B.; Pawelec, G.; Leng, S.X. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res. Rev. 2021, 65, 101205. [Google Scholar] [CrossRef]
- Bajaj, V.; Gadi, N.; Spihlman, A.P.; Wu, S.C.; Choi, C.H.; Moulton, V.R. Aging, Immunity, and COVID-19: How Age Influences the Host Immune Response to Coronavirus Infections? Front. Physiol. 2021, 11, 571416. [Google Scholar] [CrossRef] [PubMed]
- Bernadou, A.; Bouges, S.; Catroux, M.; Rigaux, J.C.; Laland, C.; Levêque, N.; Noury, U.; Larrieu, S.; Acef, S.; Habold, D.; et al. High impact of COVID-19 outbreak in a nursing home in the Nouvelle-Aquitaine region, France, March to April 2020. BMC Infect. Dis. 2021, 21, 1–6. [Google Scholar] [CrossRef]
- Kunz, R.; Minder, M. COVID-19 pandemic: Palliative care for elderly and frail patients at home and in residential and nursing homes. Swiss Med. Wkly. 2020, 150, w20235. [Google Scholar] [CrossRef] [PubMed]
- Fallon, A.; Dukelow, T.; Kennelly, S.P.; O’Neill, D. COVID-19 in nursing homes. QJM Int. J. Med. 2020, 113, 391–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrams, H.R.; Loomer, L.; Gandhi, A.; Grabowski, D.C. Characteristics of U.S. Nursing Homes with COVID-19 Cases. J. Am. Geriatr. Soc. 2020, 68, 1653–1656. [Google Scholar] [CrossRef]
- Grabenhorst, U.; Stiels-Prechtel, R.; Niemann, M.; Weckbecker, K. COVID-19 in the nursing home: A case report. MMW Fortschr. Med. 2020, 162, 60–62. [Google Scholar] [CrossRef]
- Gunst, J.D.; Staerke, N.B.; Pahus, M.H.; Kristensen, L.H.; Bodilsen, J.; Lohse, N.; Dalgaard, L.S.; Brønnum, D.; Fröbert, O.; Hønge, B.; et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with COVID-19-a double-blind randomized controlled trial. EClinicalMedicine 2021, 35, 100849. [Google Scholar] [CrossRef]
- Schutte, A.E.; Harrison, D.G. Immunity, inflammation and the vasculature in the COVID-19 era. J. Hypertens. 2020, 38, 1701–1702. [Google Scholar] [CrossRef]
- Mohammad, S.; Aziz, R.; Al Mahri, S.; Malik, S.S.; Haji, E.; Khan, A.H.; Khatlani, T.S.; Bouchama, A. Obesity and COVID-19: What makes obese host so vulnerable? Immun. Ageing 2021, 18, 1–10. [Google Scholar] [CrossRef]
- Bae, S.; Kim, S.R.; Kim, M.N.; Shim, W.J.; Park, S.M. Impact of cardiovascular disease and risk factors on fatal outcomes in patients with COVID-19 according to age: A systematic review and meta-analysis. Heart 2021, 107, 373–380. [Google Scholar] [CrossRef]
- Kim, W.-K.; McGary, C.M.; Holder, G.E.; Filipowicz, A.R.; Kim, M.M.; Beydoun, H.A.; Cai, Y.; Liu, X.; Sugimoto, C.; Kuroda, M.J. Increased Expression of CD169 on Blood Monocytes and Its Regulation by Virus and CD8 T Cells in Macaque Models of HIV Infection and AIDS. AIDS Res. Hum. Retrovir. 2015, 31, 696–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedin, A.-S.; Makinson, A.; Picot, M.-C.; Mennechet, F.; Malergue, F.; Pisoni, A.; Nyiramigisha, E.; Montagnier, L.; Bollore, K.; Debiesse, S.; et al. Monocyte CD169 Expression as a Biomarker in the Early Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2021, 223, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Tal, M.C.; Dulgeroff, L.B.T.; Myers, L.; Cham, L.B.; Mayer-Barber, K.D.; Bohrer, A.C.; Castro, E.; Yiu, Y.Y.; Angel, C.L.; Pham, E.; et al. Upregulation of CD47 Is a Host Checkpoint Response to Pathogen Recognition. mBio 2020, 11, e01293-20. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Liu, C.; Su, L.; Zhang, D.; Fan, J.; Yang, Y.; Xiao, M.; Xie, J.; Xu, Y.; et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020, 26, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Borges, R.C.; Hohmann, M.S.; Borghi, S.M. Dendritic cells in COVID-19 immunopathogenesis: Insights for a possible role in determining disease outcome. Int. Rev. Immunol. 2021, 40, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Price, J.D.; Tarbell, K.V. The Role of Dendritic Cell Subsets and Innate Immunity in the Pathogenesis of Type 1 Diabetes and Other Autoimmune Diseases. Front. Immunol. 2015, 6, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Eeden, C.; Khan, L.; Osman, M.S.; Cohen Tervaert, J.W. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int. J. Mol. Sci. 2020, 21, 6351. [Google Scholar] [CrossRef]
- Frank, K.; Paust, S. Dynamic Natural Killer Cell and T Cell Responses to Influenza Infection. Front. Cell. Infect. Microbiol. 2020, 10, 425. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, Z.; Wei, H. Developmental and Functional Control of Natural Killer Cells by Cytokines. Front. Immunol. 2017, 8, 930. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Santa Cruz, A.; Mendes-Frias, A.; Oliveira, A.I.; Dias, L.; Matos, A.R.; Carvalho, A.; Capela, C.; Pedrosa, J.; Castro, A.G.; Silvestre, R. Interleukin-6 Is a Biomarker for the Development of Fatal Severe Acute Respiratory Syndrome Coronavirus 2 Pneumonia. Front. Immunol. 2021, 12, 613422. [Google Scholar] [CrossRef]
- Ali, N. Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J. Med. Virol. 2020, 92, 2409–2411. [Google Scholar] [CrossRef] [PubMed]
- Carubbi, F.; Salvati, L.; Alunno, A.; Maggi, F.; Borghi, E.; Mariani, R.; Mai, F.; Paoloni, M.; Ferri, C.; Desideri, G.; et al. Ferritin is associated with the severity of lung involvement but not with worse prognosis in patients with COVID-19: Data from two Italian COVID-19 units. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ahnach, M.; Zbiri, S.; Nejjari, S.; Ousti, F.; Elkettani, C. C-reactive protein as an early predictor of COVID-19 severity. J. Med. Biochem. 2020, 39, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Sabaka, P.; Koščálová, A.; Straka, I.; Hodosy, J.; Lipták, R.; Kmotorková, B.; Kachlíková, M.; Kušnírová, A. Role of interleukin 6 as a predictive factor for a severe course of COVID-19: Retrospective data analysis of patients from a long-term care facility during COVID-19 outbreak. BMC Infect. Dis. 2021, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, e1008737. [Google Scholar] [CrossRef]
- Koutsakos, M.; McWilliam, H.E.; Aktepe, T.E.; Fritzlar, S.; Illing, P.; Mifsud, N.; Purcell, A.; Rockman, S.; Reading, P.C.; Vivian, J.P.; et al. Downregulation of MHC Class I Expression by Influenza A and B Viruses. Front. Immunol. 2019, 10, 1158. [Google Scholar] [CrossRef]
- Moser, E.; Hufford, M.M.; Braciale, T.J. Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3+ Regulatory T Cell Dependent Manner. PLOS Pathog. 2014, 10, e1004315. [Google Scholar] [CrossRef] [Green Version]
- Tjan, L.H.; Furukawa, K.; Nagano, T.; Kiriu, T.; Nishimura, M.; Arii, J.; Hino, Y.; Iwata, S.; Nishimura, Y.; Mori, Y. Early Differences in Cytokine Production by Severity of Coronavirus Disease 2019. J. Infect. Dis. 2021, 223, 1145–1149. [Google Scholar] [CrossRef]
- Bhat, P.; Leggatt, G.; Waterhouse, N.; Frazer, I.H. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017, 8, e2836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Kusnadi, A.; Ramirez-Suastegui, C.; Fajardo, V.; Chee, S.J.; Meckiff, B.J.; Simon, H.; Pelosi, E.; Seumois, G.; Ay, F.; Vijayanand, P.; et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021, 6, eabe4782. [Google Scholar] [CrossRef] [PubMed]
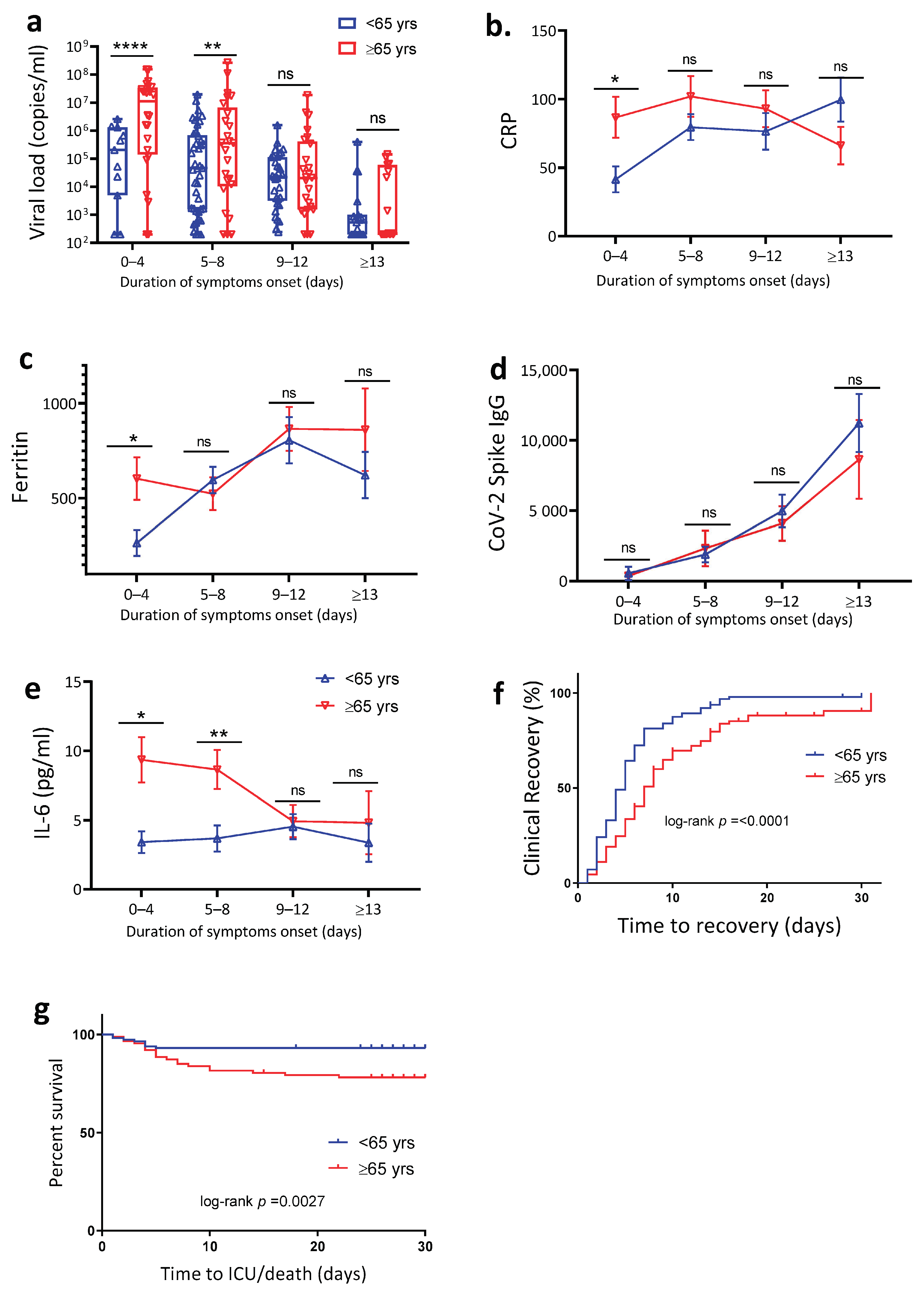


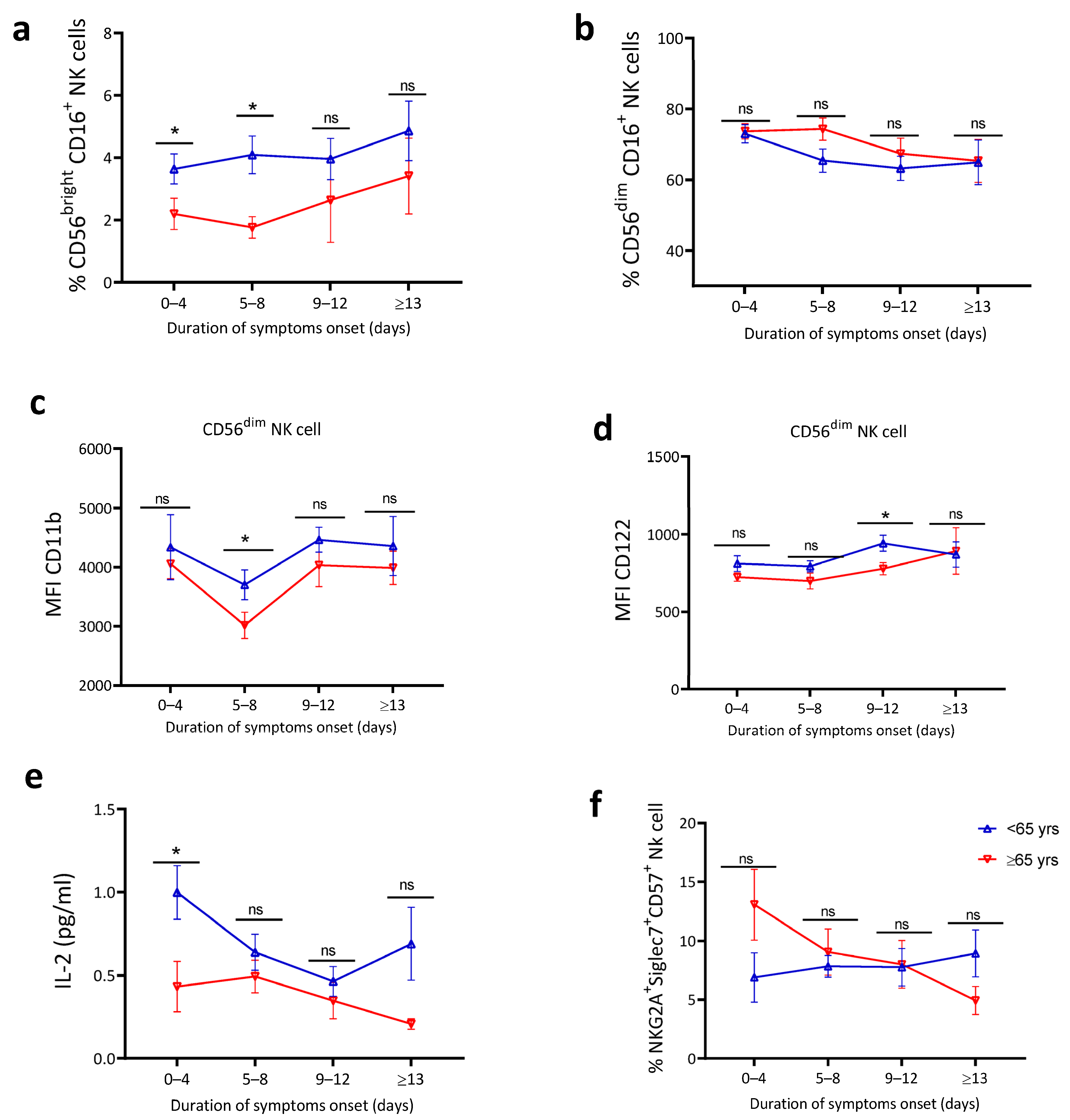
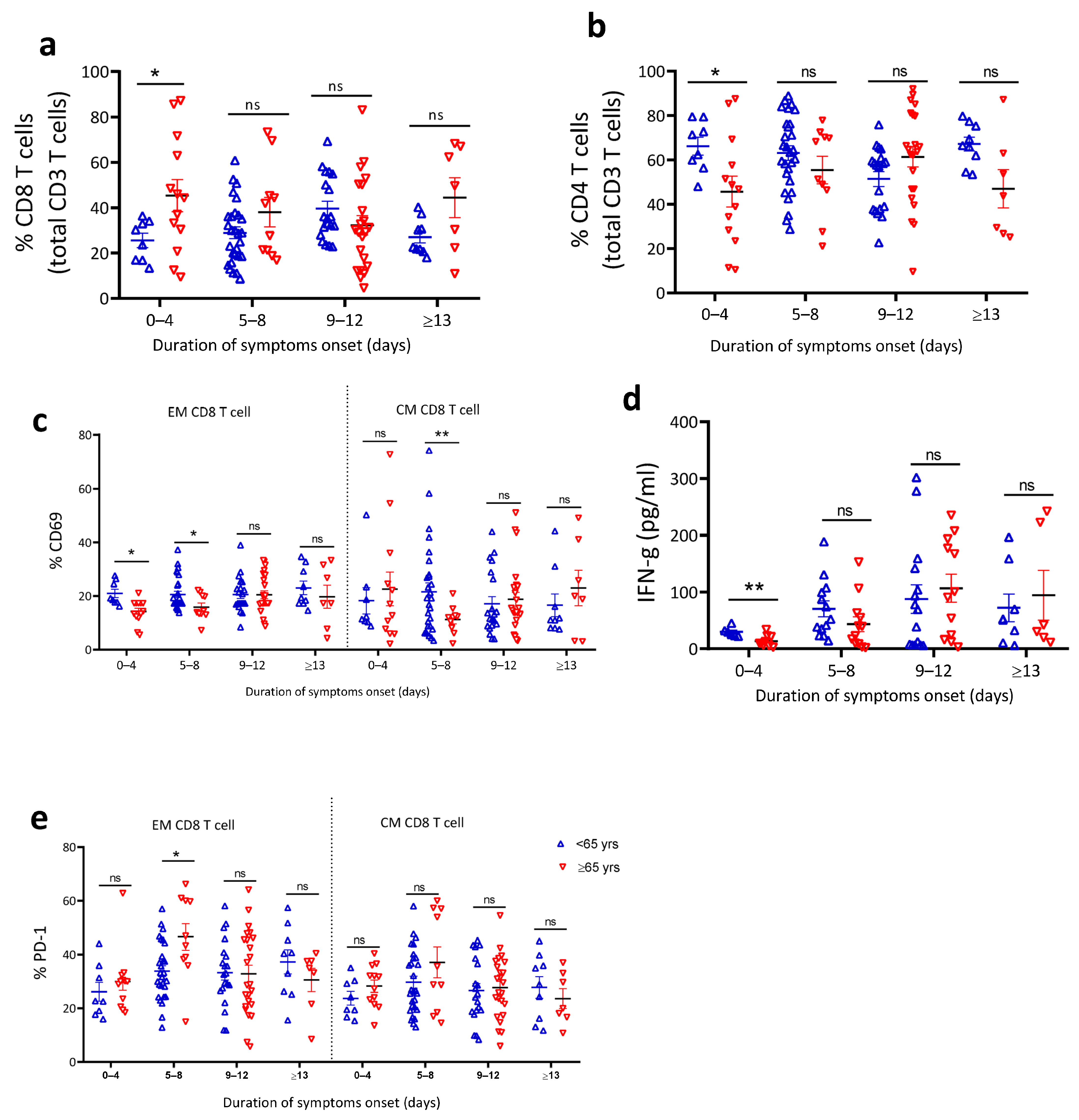
| Characteristics | <65 Years of Age (n = 116) | ≥65 Years of Age (n = 89) | p-Value |
|---|---|---|---|
| Median age (IQR)—years | 53 (46–59) | 75 (71–80) | <0.005 |
| Male sex—no. (%) | 65 (56) | 58 (65) | 0.20 |
| Median time (IQR) from symptom onset to baseline—days | 9.0 (6.0–12.0) | 7.0 (4.0–11.0) | 0.01 |
| Median weight (IQR)—kg ¥ | 90 (76–100) | 83 (72–92) | 0.01 |
| Median body–mass index (IQR)—kg/m2 ¤ | 28.3 (26.1–32.7) | 26.6 (23.3–31.4) | 0.01 |
| Obesity—no. (%) § | 44 (38) | 23 (26) | 0.07 |
| Symptoms—no. (%) | |||
| Cough | 102 (88) | 72 (81) | 0.17 |
| Dyspnea | 86 (74) | 51 (57) | 0.02 |
| Fatigue | 105 (91) | 76 (85) | 0.28 |
| Headache | 75 (65) | 32 (36) | <0.005 |
| Coexisting conditions—no. (%) | |||
| Asthma | 17 (15) | 10 (11) | 0.54 |
| COPD | 6 (05) | 15 (17) | <0.005 |
| Coronary heart disease | 10 (09) | 29 (33) | <0.005 |
| Hypertension | 27 (23) | 44 (49) | <0.005 |
| Malignancy | 4 (03) | 25 (28) | <0.005 |
| Type 2 diabetes | 15 (13) | 20 (22) | 0.09 |
| Score on 7–point ordinal scale—no. (%) | |||
| 3. Hospitalized, not requiring supplemental oxygen, requiring ongoing medical care | 48 (41) | 21 (24) | 0.03 |
| 4. Hospitalized, requiring supplemental oxygen | 60 (52) | 60 (67) | |
| 5. Hospitalized, requiring high-flow oxygen therapy or noninvasive ventilation National Early Warning Score 2—median (IQR) | 8 (07) 4 (2–6) | 8 (09) 5 (3–6) | 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cham, L.B.; Pahus, M.H.; Grønhøj, K.; Olesen, R.; Ngo, H.; Monrad, I.; Kjolby, M.; Tolstrup, M.; Gunst, J.D.; Søgaard, O.S. Effect of Age on Innate and Adaptive Immunity in Hospitalized COVID-19 Patients. J. Clin. Med. 2021, 10, 4798. https://doi.org/10.3390/jcm10204798
Cham LB, Pahus MH, Grønhøj K, Olesen R, Ngo H, Monrad I, Kjolby M, Tolstrup M, Gunst JD, Søgaard OS. Effect of Age on Innate and Adaptive Immunity in Hospitalized COVID-19 Patients. Journal of Clinical Medicine. 2021; 10(20):4798. https://doi.org/10.3390/jcm10204798
Chicago/Turabian StyleCham, Lamin B., Marie Høst Pahus, Kristoffer Grønhøj, Rikke Olesen, Hien Ngo, Ida Monrad, Mads Kjolby, Martin Tolstrup, Jesper Damsgaard Gunst, and Ole S. Søgaard. 2021. "Effect of Age on Innate and Adaptive Immunity in Hospitalized COVID-19 Patients" Journal of Clinical Medicine 10, no. 20: 4798. https://doi.org/10.3390/jcm10204798
APA StyleCham, L. B., Pahus, M. H., Grønhøj, K., Olesen, R., Ngo, H., Monrad, I., Kjolby, M., Tolstrup, M., Gunst, J. D., & Søgaard, O. S. (2021). Effect of Age on Innate and Adaptive Immunity in Hospitalized COVID-19 Patients. Journal of Clinical Medicine, 10(20), 4798. https://doi.org/10.3390/jcm10204798






