Machine Learning Refutes Loss of Smell as a Risk Indicator of Diabetes Mellitus
Abstract
:1. Introduction
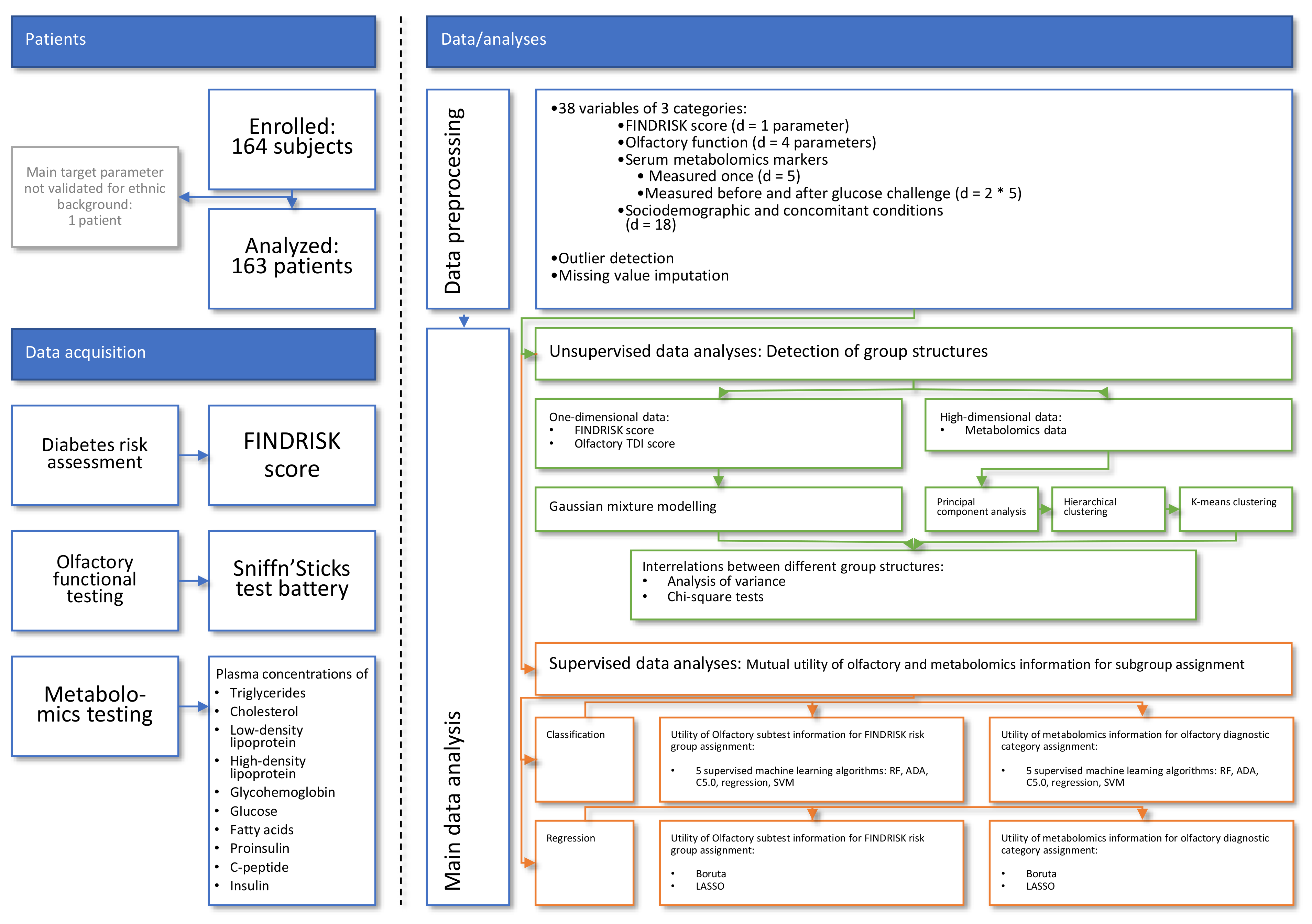
2. Materials and Methods
2.1. Study Design and Participants
2.2. Data Acquisition
2.2.1. Diabetes Risk Assessment
2.2.2. Clinical Testing of Olfactory Function
2.2.3. Metabolomics Testing
2.3. Data Analysis
2.3.1. Data Prepossessing
2.3.2. Detection of Group Structures in Metabolomic and Olfactory Data
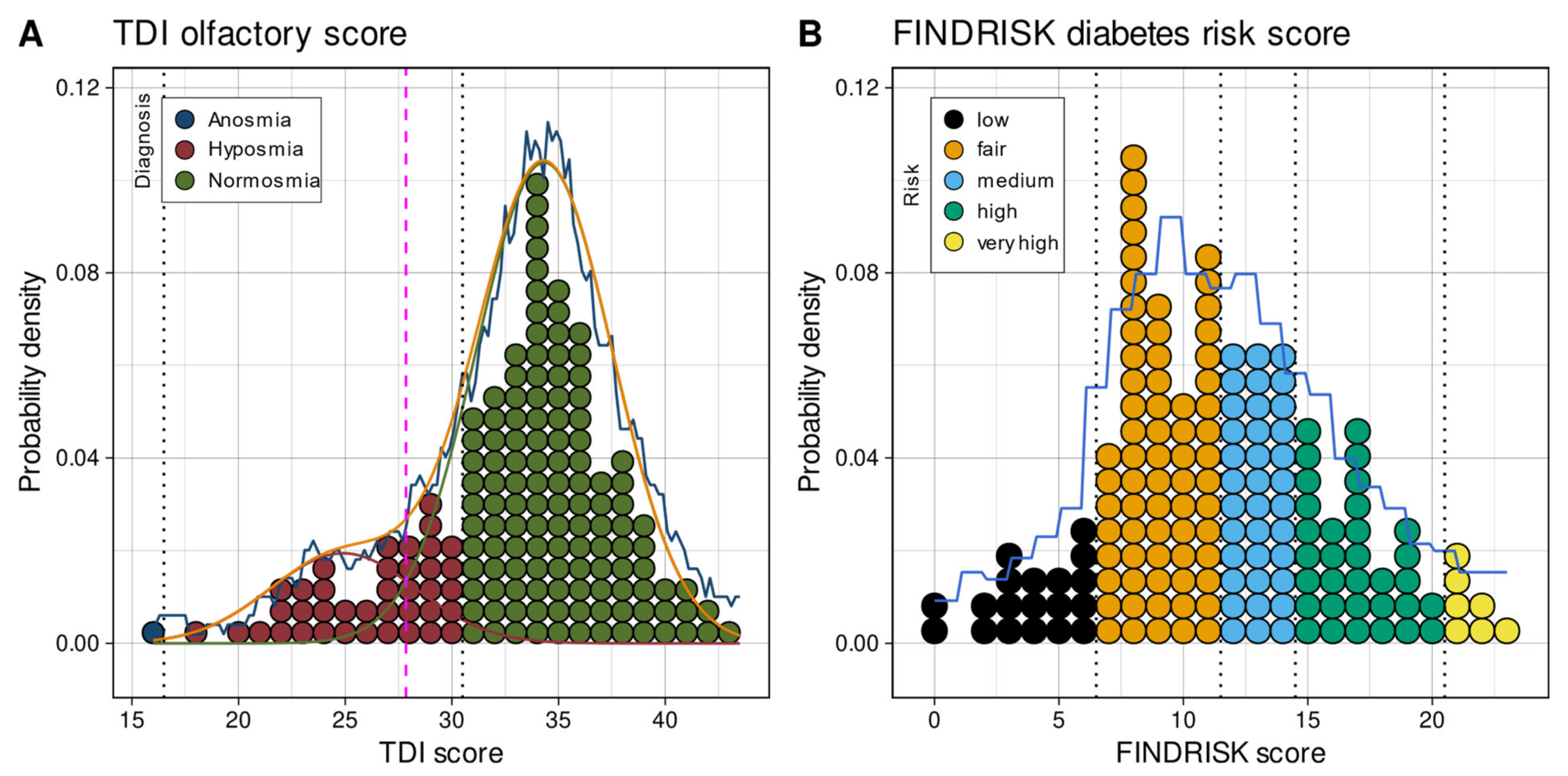
Assessment of Group Structures in One-Dimensional Olfactory and Diabetes Risk Data
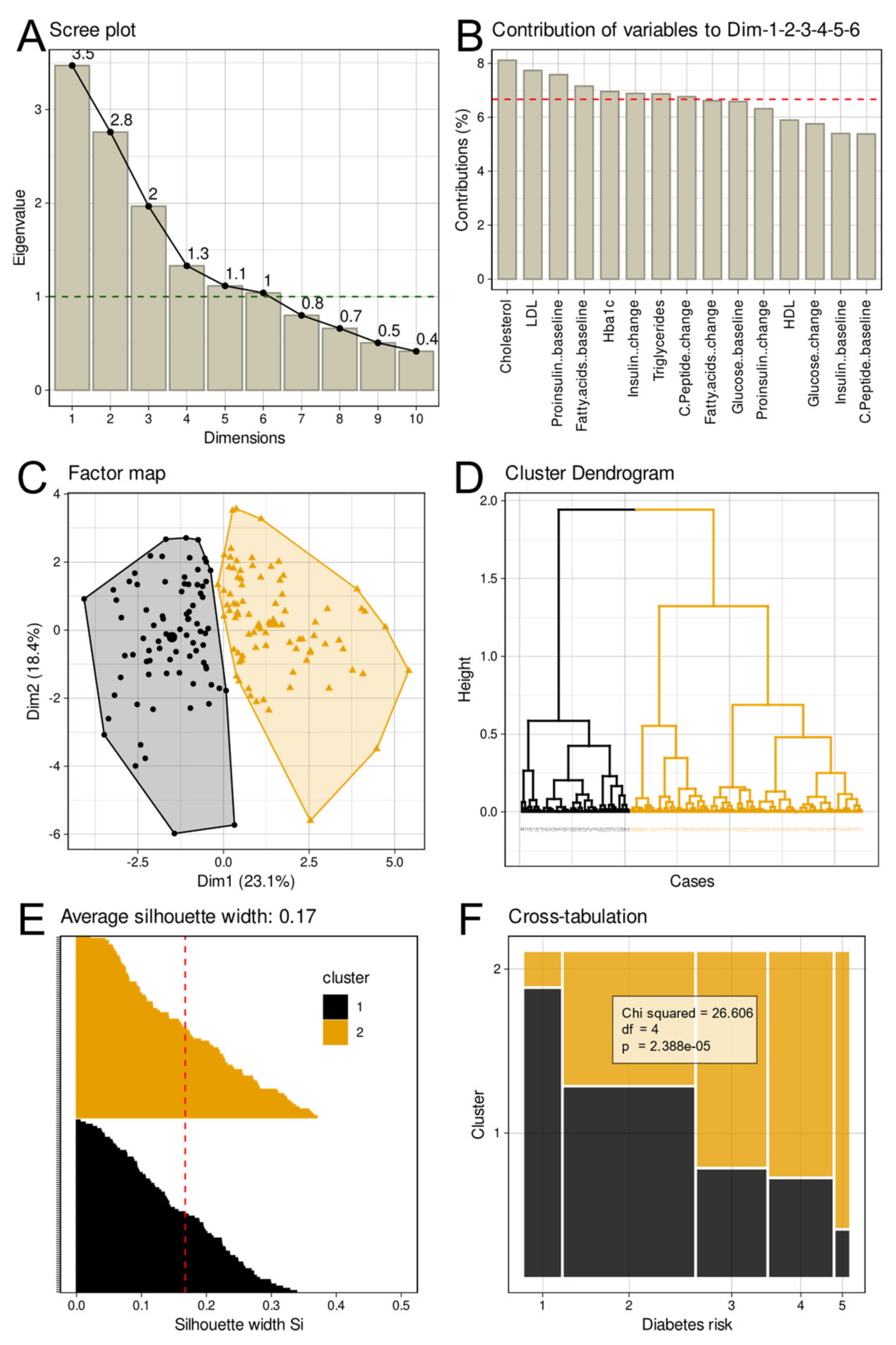
Assessment of Group Structures in High-Dimensional Metabolomics Data
2.3.3. Investigation of Interrelations between Different Group Structures
Statistical Analysis of the Association between Odor Information and Diabetes Risk
Evaluation of the Utility of Olfactory and Metabolomic Information in Predicting Diabetes Risk
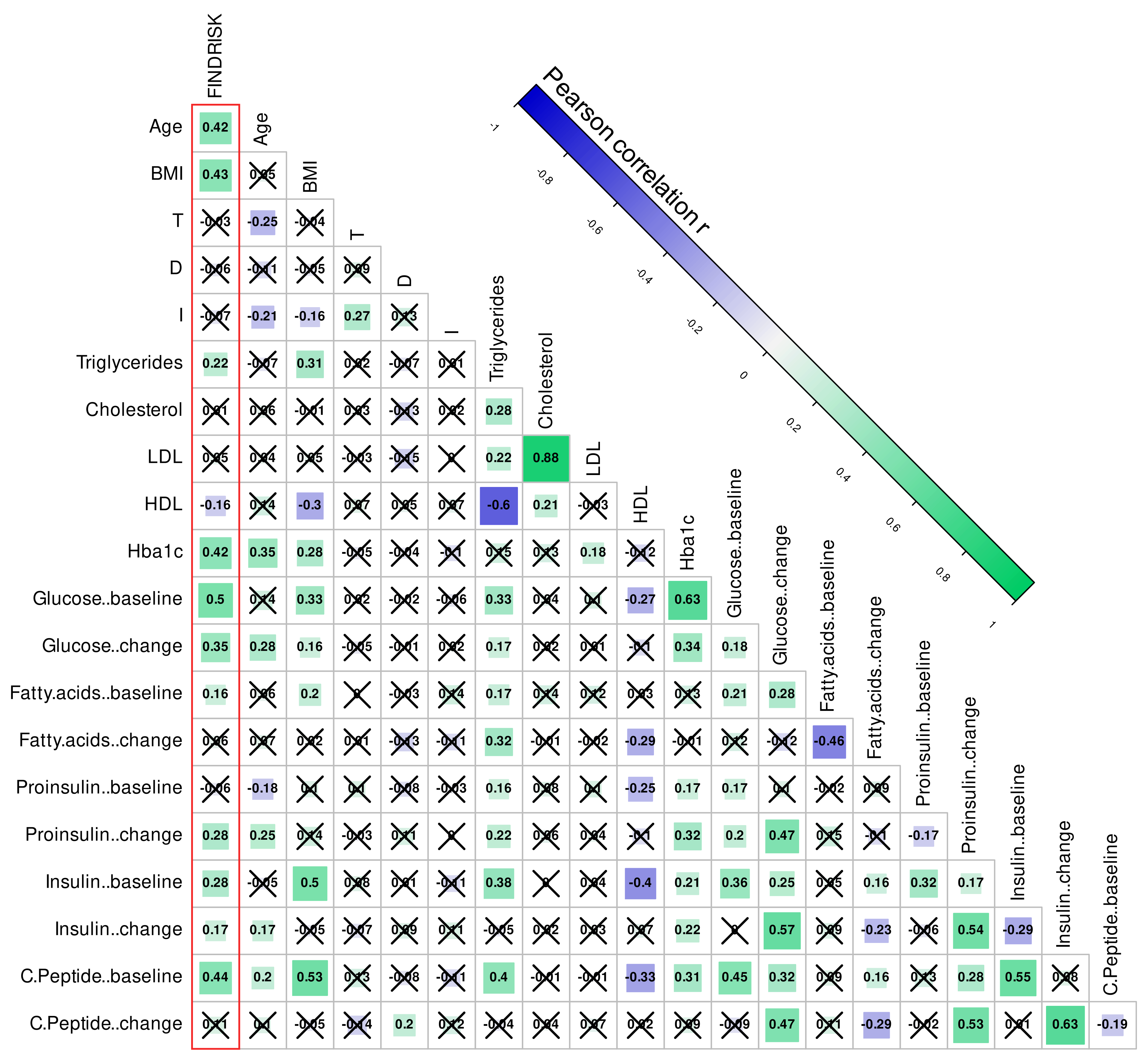
2.3.4. Exploration of the Associations of Potential Confounders with Diabetes Risk
3. Results
3.1. One- and High-Dimensional Group Structures in Metabolomic and Olfactory Data
3.2. Interrelationships between Different Group Structures
3.2.1. Results of Statistical Analyses of the Association between Olfactory Information and Diabetes Risk
3.2.2. Utility of Olfactory and Metabolomic Information in Predicting Diabetes Risk
Machine-Learned Classification Approach
Machine-Learned Regression Approach
3.3. Associations of Medical or Other Factors or Potential Confounders with Diabetes Risk
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doty, R.L. Clinical disorders of olfaction. Handb. Olfaction Gustation 2015, 375–402. [Google Scholar] [CrossRef]
- Upadhyay, U.D.; Holbrook, E.H. Olfactory loss as a result of toxic exposure. Otolaryngol. Clin. North Am. 2004, 37, 1185–1207. [Google Scholar] [CrossRef] [PubMed]
- Klopfenstein, T.; Kadiane-Oussou, N.J.; Toko, L.; Royer, P.Y.; Lepiller, Q.; Gendrin, V.; Zayet, S. Features of anosmia in COVID-19. Med. Mal. Infect. 2020, 50, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Hummel, T.; Whitcroft, K.L.; Andrews, P.; Altundag, A.; Cinghi, C.; Costanzo, R.M.; Damm, M.; Frasnelli, J.; Gudziol, H.; Gupta, N.; et al. Position paper on olfactory dysfunction. Rhinology. Suppl. 2017, 54, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Ansari, K.A.; Johnson, A. Olfactory function in patients with Parkinson’s disease. J. Chron. Dis. 1975, 28, 493–497. [Google Scholar] [CrossRef]
- Doty, R.L.; Deems, D.; Steller, S. Olfactory dysfunction in Parkinson’s disease: A general deficit unrelated to neurologic signs, disease stage, or disease duration. Neurology 1988, 38, 1237–1244. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, P.; Schuurman, T.; Prinz, H. Loss of smell leads to dementia in mice: Is Alzheimer’s disease a degenerative disorder of the olfactory system? J. Protein Chem. 1989, 8, 448–451. [Google Scholar] [CrossRef]
- Hawkes, C.H. Assessment of olfaction in multiple sclerosis. Chem. Senses 1996, 21, 486. [Google Scholar]
- Graham, C.S.; Graham, B.G.; Bartlett, J.A.; Heald, A.E.; Schiffman, S.S. Taste and smell losses in HIV infected patients. Physiol. Behav. 1995, 58, 287–293. [Google Scholar] [CrossRef]
- Haehner, A.; Hummel, T.; Hummel, C.; Sommer, U.; Junghanns, S.; Reichmann, H. Olfactory loss may be a first sign of idiopathic Parkinson’s disease. Mov. Disord. 2007, 22, 839–842. [Google Scholar] [CrossRef]
- Haehner, A.; Masala, C.; Walter, S.; Reichmann, H.; Hummel, T. Incidence of Parkinson’s disease in a large patient cohort with idiopathic smell and taste loss. J. Neurol. 2019, 266, 339–345. [Google Scholar] [CrossRef]
- Campabadal, A.; Uribe, C.; Segura, B.; Baggio, H.C.; Abos, A.; Garcia-Diaz, A.I.; Marti, M.J.; Valldeoriola, F.; Compta, Y.; Bargallo, N.; et al. Brain correlates of progressive olfactory loss in Parkinson’s disease. Parkinsonism Relat. Disord. 2017, 41, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Chen, B.; Zhong, X.; Zhou, H.; Zhang, M.; Mai, N.; Wu, Z.; Huang, X.; Haehner, A.; Chen, X.; et al. Olfactory dysfunction is already present with subjective cognitive decline and deepens with disease severity in the Alzheimer’s disease spectrum. J. Alzheimer’s Dis. 2021, 79, 585–595. [Google Scholar] [CrossRef]
- Rasmussen, V.F.; Vestergaard, E.T.; Hejlesen, O.; Andersson, C.U.N.; Cichosz, S.L. Prevalence of taste and smell impairment in adults with diabetes: A cross-sectional analysis of data from the National Health and Nutrition Examination Survey (NHANES). Prim. Care Diabetes 2018, 12, 453–459. [Google Scholar] [CrossRef]
- Chan, J.Y.K.; García-Esquinas, E.; Ko, O.H.; Tong, M.C.F.; Lin, S.Y. The association between diabetes and olfactory function in adults. Chem. Senses 2017, 43, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Zaghloul, H.; Pallayova, M.; Al-Nuaimi, O.; Hovis, K.R.; Taheri, S. Association between diabetes mellitus and olfactory dysfunction: Current perspectives and future directions. Diabet. Med. 2018, 35, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Weinstock, R.S.; Wright, H.N.; Smith, D.U. Olfactory dysfunction in diabetes mellitus. Physiol. Behav. 1993, 53, 17–21. [Google Scholar] [CrossRef]
- Takayama, S.; Sasaki, T. Acute hyposmia in type 2 diabetes. J. Int. Med. Res. 2003, 31, 466–468. [Google Scholar] [CrossRef]
- Heckmann, J.G.; Höcherl, C.; Dütsch, M.; Lang, C.; Schwab, S.; Hummel, T. Smell and taste disorders in polyneuropathy: A prospective study of chemosensory disorders. Acta Neurol. Scand. 2009, 120, 258–263. [Google Scholar] [CrossRef]
- Guthoff, M.; Tschritter, O.; Berg, D.; Liepelt, I.; Schulte, C.; Machicao, F.; Haering, H.U.; Fritsche, A. Effect of genetic variation in Kv1.3 on olfactory function. Diabetes Metab. Res. Rev. 2009, 25, 523–527. [Google Scholar] [CrossRef]
- Le Floch, J.P.; Le Lièvre, G.; Labroue, M.; Paul, M.; Peynegre, R.; Perlemuter, L. Smell dysfunction and related factors in diabetic patients. Diabetes Care 1993, 16, 934–937. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, B.; Wang, X.; Zhang, X.; Yang, Q.X.; Qing, Z.; Zhang, W.; Zhu, D.; Bi, Y. Olfactory dysfunction mediates adiposity in cognitive impairment of type 2 diabetes: Insights from clinical and functional neuroimaging studies. Diabetes Care 2019, 42, 1274–1283. [Google Scholar] [CrossRef]
- Gouveri, E.; Katotomichelakis, M.; Gouveris, H.; Danielides, V.; Maltezos, E.; Papanas, N. Olfactory dysfunction in type 2 diabetes mellitus: An additional manifestation of microvascular disease? Angiology 2014, 65, 869–876. [Google Scholar] [CrossRef]
- Brady, S.; Lalli, P.; Midha, N.; Chan, A.; Garven, A.; Chan, C.; Toth, C. Presence of neuropathic pain may explain poor performances on olfactory testing in diabetes mellitus patients. Chem. Senses 2013, 38, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Gascón, C.; Santaolalla, F.; Martínez, A.; Sánchez Del Rey, A. Usefulness of the BAST-24 smell and taste test in the study of diabetic patients: A new approach to the determination of renal function. Acta Otolaryngol. 2013, 133, 400–404. [Google Scholar] [CrossRef]
- Sanke, H.; Mita, T.; Yoshii, H.; Someya, Y.; Yamashiro, K.; Shimizu, T.; Ohmura, C.; Onuma, T.; Watada, H. Olfactory dysfunction predicts the development of dementia in older patients with type 2 diabetes. Diabetes Res. Clin. Pract. 2021, 174, 108740. [Google Scholar] [CrossRef]
- Doty, R.L.; Stern, M.B.; Pfeiffer, C.; Gollomp, S.M.; Hurtig, H.I. Bilateral olfactory dysfunction in early stage treated and untreated idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1992, 55, 138–142. [Google Scholar] [CrossRef]
- Constantinescu, C.S.; Raps, E.C.; Cohen, J.A.; West, S.E.; Doty, R.L. Olfactory disturbances as the initial or most prominent symptom of multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 1994, 57, 1011–1012. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, B.; Wang, X.; Zhang, X.; Yang, Q.X.; Qing, Z.; Lu, J.; Bi, Y.; Zhu, D. Altered odor-induced brain activity as an early manifestation of cognitive decline in patients with type 2 diabetes. Diabetes 2018, 67, 994–1006. [Google Scholar] [CrossRef] [Green Version]
- Rajput, D.S.; Basha, S.M.; Xin, Q.; Gadekallu, T.R.; Kaluri, R.; Lakshmanna, K.; Maddikunta, P.K.R. Providing diagnosis on diabetes using cloud computing environment to the people living in rural areas of India. J. Ambient. Intell. Humaniz. Comput. 2021. [Google Scholar] [CrossRef]
- Herman, W.H.; Ye, W.; Griffin, S.J.; Simmons, R.K.; Davies, M.J.; Khunti, K.; Rutten, G.E.H.M.; Sandbaek, A.; Lauritzen, T.; Borch-Johnsen, K.; et al. Early detection and treatment of type 2 diabetes reduce cardiovascular morbidity and mortality: A simulation of the results of the anglo-danish-dutch study of intensive treatment in people with screen-detected diabetes in primary care (ADDITION-Europe). Diabetes Care 2015, 38, 1449–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindström, J.; Tuomilehto, J. The diabetes risk score: A practical tool to predict type 2 diabetes risk. Diabetes Care 2003, 26, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Thorand, B.; Osterhoff, M.A.; Muller, G.; Bohm, A.; Meisinger, C.; Kowall, B.; Rathmann, W.; Kronenberg, F.; Staiger, H.; et al. Family history of diabetes is associated with higher risk for prediabetes: A multicentre analysis from the German Center for Diabetes Research. Diabetologia 2013, 56, 2176–2180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, P.E.H.; Li, J.; Lindstrom, J.; Tuomilehto, J. Tools for predicting the risk of type 2 diabetes in daily practice. Horm. Metab. Res. 2009, 41, 86–97. [Google Scholar] [CrossRef]
- De Feo, P.; Schwarz, P. Is physical exercise a core therapeutical element for most patients with type 2 diabetes? Diabetes Care 2013, 36, S149–S154. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.; Allison, D.B.; Heymsfield, S.B.; Kelley, D.E.; Leibel, R.L.; Nonas, C.; Kahn, R.; Association for Weight Management and Obesity Prevention; NAASO; Obesity Society; et al. Waist circumference and cardiometabolic risk: A consensus statement from shaping America’s health: Association for Weight Management and Obesity Prevention; NAASO, the Obesity Society; the American Society for Nutrition; and the American Diabetes Association. Diabetes Care 2007, 30, 1647–1652. [Google Scholar] [CrossRef] [Green Version]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Buse, J.B.; Wexler, D.J.; Tsapas, A.; Rossing, P.; Mingrone, G.; Mathieu, C.; D’Alessio, D.A.; Davies, M.J. 2019 Update to: Management of hyperglycemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care 2020, 43, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Thamer, C.; Machann, J.; Stefan, N.; Haap, M.; Schafer, S.; Brenner, S.; Kantartzis, K.; Claussen, C.; Schick, F.; Haring, H.; et al. High visceral fat mass and high liver fat are associated with resistance to lifestyle intervention. Obesity 2007, 15, 531–538. [Google Scholar] [CrossRef]
- Ji, Y.; Yiorkas, A.M.; Frau, F.; Mook-Kanamori, D.; Staiger, H.; Thomas, E.L.; Atabaki-Pasdar, N.; Campbell, A.; Tyrrell, J.; Jones, S.E.; et al. Genome-wide and abdominal MRI data provide evidence that a genetically determined favorable adiposity phenotype is characterized by lower ectopic liver fat and lower risk of type 2 diabetes, heart disease, and hypertension. Diabetes 2019, 68, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Barr, E.L.; Cameron, A.J.; Balkau, B.; Zimmet, P.Z.; Welborn, T.A.; Tonkin, A.M.; Shaw, J.E. HOMA insulin sensitivity index and the risk of all-cause mortality and cardiovascular disease events in the general population: The Australian Diabetes, Obesity and Lifestyle Study (AusDiab) study. Diabetologia 2010, 53, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Heianza, Y.; Hara, S.; Arase, Y.; Saito, K.; Fujiwara, K.; Tsuji, H.; Kodama, S.; Hsieh, S.D.; Mori, Y.; Shimano, H.; et al. HbA1c 5.7–6.4% and impaired fasting plasma glucose for diagnosis of prediabetes and risk of progression to diabetes in Japan (TOPICS 3): A longitudinal cohort study. Lancet 2011, 378, 147–155. [Google Scholar] [CrossRef]
- Clark, C.M.; Fradkin, J.E.; Hiss, R.G.; Lorenz, R.A.; Vinicor, F.; Warren-Boulton, E. Promoting early diagnosis and treatment of type 2 diabetesthe national diabetes education program. JAMA 2000, 284, 363–365. [Google Scholar] [CrossRef]
- Lötsch, J.; Geisslinger, G.; Hummel, T. Sniffing out pharmacology: Interactions of drugs with human olfaction. Trends Pharmacol. Sci. 2012, 33, 193–199. [Google Scholar] [CrossRef]
- Lötsch, J.; Daiker, H.; Hähner, A.; Ultsch, A.; Hummel, T. Drug-target based cross-sectional analysis of olfactory drug effects. Eur. J. Clin. Pharmacol. 2015, 71, 461–471. [Google Scholar] [CrossRef]
- Schwarz, P. FINDRISK—Test für Diabetesrisiko. Der Diabetol. 2020, 16, 524–526. [Google Scholar] [CrossRef]
- Hummel, T.; Hummel, C.; Welge-Luessen, A. Assessment of olfaction and gustation. In Management of Smell and Taste Disorders—A Practical Guide for Clinicians; Welge-Luessen, A., Hummel, T., Eds.; Thieme: Stuttgart, Germany, 2013; pp. 58–75. [Google Scholar]
- Kobal, G.; Hummel, T.; Sekinger, B.; Barz, S.; Roscher, S.; Wolf, S.R. “Sniffin’ sticks”: Screening of olfactory performance. Rhinology 1996, 34, 222–226. [Google Scholar]
- Hummel, T.; Sekinger, B.; Wolf, S.R.; Pauli, E.; Kobal, G. ‘Sniffin’ sticks’: Olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chem. Senses 1997, 22, 39–52. [Google Scholar] [CrossRef]
- Oleszkiewicz, A.; Schriever, V.A.; Croy, I.; Hahner, A.; Hummel, T. Updated sniffin’ sticks normative data based on an extended sample of 9139 subjects. Eur. Arch. Oto Rhino Laryngol. 2019, 276, 719–728. [Google Scholar] [CrossRef] [Green Version]
- Cefalu, W.T.; Berg, E.G.; Saraco, M.; Petersen, M.P.; Uelmen, S.; Robinson, S. Classification and diagnosis of diabetes: Standards of medical care in diabetes—2019. Diabetes Care 2019, 42, S13–S28. [Google Scholar] [CrossRef] [Green Version]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 498–520. [Google Scholar] [CrossRef]
- Pearson, K.; Lines, L.O. Planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Smirnov, N. Table for estimating the goodness of fit of empirical distributions. Ann. Math. Stat. 1948, 19, 279–281. [Google Scholar] [CrossRef]
- Grubbs, F.E. Sample Criteria for Testing Outlying Observations. Ann. Math. Statist. 1950, 21, 27–58. [Google Scholar] [CrossRef]
- Komsta, L. Outliers: Tests for outliers. 2011. [Google Scholar]
- Torgo, L. Data Mining Using R: Learning with Case Studies; Chapman & Hall: London, UK, 2010. [Google Scholar]
- Ultsch, A. Pareto density estimation: A density estimation for knowledge discovery. In Innovations in Classification, Data Science, and Information Systems, Proceedings of the 27th Annual Conference of the German Classification Society (GfKL), Technische Universität Cottbus, Germany, 12–14 March 2003; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Bayes, T. An essay towards solving a problem in the doctrine of chances. By the Late Rev. Mr. Bayes, F. R. S. Communicated by Mr. Price, in a Letter to John Canton, A. M. F. R. S. Philos. Trans. 1763, 53, 370–418. [Google Scholar] [CrossRef]
- Swets, J.A. The relative operating characteristic in psychology: A technique for isolating effects of response bias finds wide use in the study of perception and cognition. Science 1973, 182, 990–1000. [Google Scholar] [CrossRef]
- Lerch, F.; Ultsch, A.; Lotsch, J. Distribution optimization: An evolutionary algorithm to separate Gaussian mixtures. Sci. Rep. 2020, 10, 648. [Google Scholar] [CrossRef] [Green Version]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, H.F. The varimax criterion for analytic rotation in factor analysis. Psychometrika 1958, 23, 187–200. [Google Scholar] [CrossRef]
- Guttman, L. Some necessary conditions for common factor analysis. Psychometrika 1954, 19, 149–161. [Google Scholar] [CrossRef]
- Kassambara, A. Practical Guide To Principal Component Methods in R: PCA, M(CA), FAMD, MFA, HCPC, Factoextra; Copyright ©2017 by Alboukadel Kassambara. Available online: http://www.sthda.com/french/ (accessed on 21 October 2021).
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June 1967; Volume 1, pp. 281–297. [Google Scholar]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions. 2017, Volume 1, p. 56. Available online: https://cran.r-project.org/package=cluster (accessed on 21 October 2021).
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. Artic. 2014, 61, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Comp. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Vavrek, M.J. fossil: Palaeoecological and palaeogeographical analysis tools. Palaeontol. Electron. 2011, 14, 1T. [Google Scholar]
- Bonferroni, C.E. Teoria statistica delle classi e calcolo delle probabilita. Pubbl. R Ist. Super. Sci. Econ. Commer. Firenze 1936, 8, 3–62, citeulike-article-id:1778138. [Google Scholar]
- Pearson, K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos. Mag. Ser. 5 1900, 50, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Doty, R.L.; Shaman, P.; Applebaum, S.L.; Giberson, R.; Siksorski, L.; Rosenberg, L. Smell identification ability: Changes with age. Science 1984, 226, 1441–1443. [Google Scholar] [CrossRef]
- Saeys, Y.; Inza, I.; Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Lee, J.S.; Paintsil, E.; Gopalakrishnan, V.; Ghebremichael, M. A comparison of machine learning techniques for classification of HIV patients with antiretroviral therapy-induced mitochondrial toxicity from those without mitochondrial toxicity. BMC Med. Res. Methodol. 2019, 19, 216. [Google Scholar] [CrossRef]
- Good, P.I. Resampling Methods: A Practical Guide to Data Analysis; Birkhäuser: Boston, MA, USA, 2006. [Google Scholar]
- Tillé, Y.; Matei, A. Sampling: Survey Sampling. 2016. Available online: https://cran.r-project.org/package=sampling (accessed on 21 October 2021).
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The balanced accuracy and its posterior distribution. In Proceedings of the 2010 20th International Conference on Pattern Recognition (ICPR), Istanbul, Turkey, 23–26 August 2010; pp. 3121–3124. [Google Scholar]
- Altman, D.G.; Bland, J.M. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; Bland, J.M. Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, T.A.A. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons; I kommission hos E. Munksgaard: København, Denmark, 1948. [Google Scholar]
- Jardine, N.; van Rijsbergen, C.J. The use of hierarchic clustering in information retrieval. Inf. Storage Retr. 1971, 7, 217–240. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and regression training. Astrophys. Source Code Libr. 2018. Available online: https://cran.r-project.org/package=caret (accessed on 21 October 2021).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, p. 278. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Schapire, R.E.; Freund, Y. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 771–780. [Google Scholar]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Berkson, J. Application of the logistic function to bio-assay. J. Am. Stat. Assoc. 1944, 39, 357–365. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting. 2020. Available online: https://cran.r-project.org/package=xgboost (accessed on 21 October 2021).
- Kuhn, M.; Quinlan, R. C50: C5.0 Decision Trees and Rule-Based Models. 2018. Available online: https://cran.r-project.org/package=C50 (accessed on 21 October 2021).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S; Springer: New York, NY, USA, 2002. [Google Scholar]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Stat. Softw. 2010, 36, 13. [Google Scholar] [CrossRef] [Green Version]
- Santosa, F.; Symes, W.W. Linear inversion of band-limited reflection seismograms. SIAM J. Sci. Stat. Comput. 1986, 7, 1307–1330. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Groemping, U. Relative importance for linear regression in R: The package relaimpo. J. Stat. Softw. 2006, 17, 27. [Google Scholar] [CrossRef] [Green Version]
- Ultsch, A.; Lötsch, J. Computed ABC analysis for rational selection of most informative variables in multivariate data. PLoS ONE 2015, 10, e0129767. [Google Scholar] [CrossRef] [Green Version]
- Lötsch, J.; Ultsch, A. Random forests followed by computed ABC analysis as a feature selection method for machine learning in biomedical data. In Advanced Studies in Classification and Data Science; Springer: Singapore, 2020; pp. 57–69. [Google Scholar]
- Juran, J.M. The non-Pareto principle; Mea culpa. Qual. Prog. 1975, 8, 8–9. [Google Scholar]
- Beard, M.D.; Mackay-Sim, A. Loss of sense of smell in adult, hypothyroid mice. Brain Res. 1987, 433, 181–189. [Google Scholar] [CrossRef]
- McConnell, R.J.; Menendez, C.E.; Smith, F.R.; Henkin, R.I.; Rivlin, R.S. Defects of taste and smell in patients with hypothyroidism. Am. J. Med. 1975, 59, 354–364. [Google Scholar] [CrossRef]
- Meyer, D.; Zeileis, A.; Hornik, K. vcd: Visualizing Categorical Data; 2016, vcd: Visualizing Categorical Data. R package version 1.4-8. Available online: https://cran.r-project.org/web/packages/vcd/citation.html (accessed on 21 October 2021).
- Student. The probable error of a mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Lötsch, J.; Knothe, C.; Lippmann, C.; Ultsch, A.; Hummel, T.; Walter, C. Olfactory drug effects approached from human-derived data. Drug Discov. Today 2015, 20, 1398–1406. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Arnold, J.B. ggthemes: Extra Themes, Scales and Geoms for ‘ggplot2′; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pearson, K. On a new method of determining the correlation between a measured character A and a character B, of which only the percentage of cases wherin B exceeds (or falls short of) a given intensity is recorded for each grade of A. Biometrika 1909, 7, 96–105. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R package “Corrplot”: Visualization of A Correlation Matrix. 2021. Available online: https://github.com/taiyun/corrplot (accessed on 21 October 2021).
- Doty, R.L.; Hawkes, C.H. Chemosensory dysfunction in neurodegenerative diseases. Handb. Clin. Neurol. 2019, 164, 325–360. [Google Scholar] [CrossRef]
- Xu, L.; Liu, J.; Wroblewski, K.E.; McClintock, M.K.; Pinto, J.M. Odor sensitivity versus odor identification in older us adults: Associations with cognition, age, gender, and race. Chem. Senses 2020, 45, 321–330. [Google Scholar] [CrossRef]
- Pinto, J.M.; Wroblewski, K.E.; Kern, D.W.; Schumm, L.P.; McClintock, M.K. Olfactory dysfunction predicts 5-year mortality in older adults. PLoS ONE 2014, 9, e107541. [Google Scholar] [CrossRef] [Green Version]
- Baskin, D.G.; Porte, D., Jr.; Guest, K.; Dorsa, D.M. Regional concentrations of insulin in the rat brain. Endocrinology 1983, 112, 898–903. [Google Scholar] [CrossRef]
- Banks, W.A.; Kastin, A.J.; Pan, W. Uptake and degradation of blood-borne insulin by the olfactory bulb. Peptides 1999, 20, 373–378. [Google Scholar] [CrossRef]
- Banks, W.A. The source of cerebral insulin. Eur. J. Pharmacol. 2004, 490, 5–12. [Google Scholar] [CrossRef]
- Lötsch, J.; Schaeffeler, E.; Mittelbronn, M.; Winter, S.; Gudziol, V.; Schwarzacher, S.W.; Hummel, T.; Doehring, A.; Schwab, M.; Ultsch, A. Functional genomics suggest neurogenesis in the adult human olfactory bulb. Brain Struct. Funct. 2014, 219, 1991–2000. [Google Scholar] [CrossRef]
- Altundag, A.; Ay, S.A.; Hira, S.; Salıhoglu, M.; Baskoy, K.; Denız, F.; Tekelı, H.; Kurt, O.; Yonem, A.; Hummel, T. Olfactory and gustatory functions in patients with non-complicated type 1 diabetes mellitus. Eur. Arch. Oto Rhino Laryngol. 2017, 274, 2621–2627. [Google Scholar] [CrossRef] [PubMed]
- Naka, A.; Riedl, M.; Luger, A.; Hummel, T.; Mueller, C.A. Clinical significance of smell and taste disorders in patients with diabetes mellitus. Eur. Arch. Oto Rhino Laryngol. 2010, 267, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Varma, S.; Simon, R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 2006, 7, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Early Sign | Method of Detection |
Key Reference |
|---|---|---|
| Genetic variation | Genetic variation | [33] |
| Family history | Relatives with diabetes | [34] |
| Age, family history, waist circumference, physical activity, consumption of vegetables etc., antihypertensive medication, blood sugar levels, body mass index | FINDRISK and similar scores | [35] |
| Physical inactivity | Time and intensity | [36] |
| Waist circumference | cm | [37] |
| Obesity | Body weight/body mass index | [38,39] |
| Visceral obesity | Fat mass/MRI | [40] |
| Liver fat | Fat mass/MRI | [41] |
| Insulin resistance | HOMA | [42] |
| Fasting hyperglycemia | glucose | [39] |
| Postprandial hyperglycemia | Oral glucose tolerance test, post meal glucose | [39] |
| HbA1c | HPLC | [39,43] |
| Waist circumference, blood pressure, mercury level, plasma triacylglycerol, blood glucose, HDL cholesterol, glucose | Support vector machines | [30] |
| Parameter | Unit | n | Range Counts/Positive Responses | Mean ± SD |
|---|---|---|---|---|
| Demographics | ||||
| Age | Years | 163 | 18–69 | 52.93 ± 12.7 |
| Sex | - | 163 | 6 men 101 women | - |
| BMI—body mass index | kg/m2 | 163 | 19.7–43.7 | 28.25 ± 5.04 |
| Waist-hip ratio | - | 163 | 0.6–1.1 | 0.92 ± 0.08 |
| Systolic blood pressure | mm Hg | 163 | 102–191 | 135.81 ± 16.91 |
| Diastolic blood pressure | mm Hg | 163 | 52–120 | 79.57 ± 11.99 |
| Prior or Concomitant Symptoms and Diseases | ||||
| Head trauma | - | 163 | 16 (9.8%) | - |
| Headache | - | 163 | 37 (22.7%) | - |
| Postnasal drip | - | 163 | 16 (9.8%) | - |
| Neurological disorder | - | 163 | 6 (3.7%) | - |
| Renal dysfunction | - | 163 | 6 (3.7%) | - |
| Nasal symptoms | - | 163 | 0 = 101 (62%) 1 = 38 (23.3%) 2 = 15 (9.2%) 3 = 6 (3.7%) 4 = 2 (1.2%) 5 = 1 (0.6%) | - |
| Snoring | - | 163 | 58 (35.6%) | - |
| Hepatitis | - | 163 | 13 (8%) | - |
| Hypothyroidism | - | 163 | 15 (9.2%) | - |
| Hyperthyroidism | - | 163 | 8 (4.9%) | - |
| Surgery: palatine tonsils | - | 163 | 20 (12.3%) | - |
| Surgery: pharyngeal tonsils | - | 163 | 11 (6.7%) | - |
| Surgery: middle ear | - | 163 | 4 (2.5%) | - |
| Surgery: teeth | - | 163 | 25 (15.3%) | - |
| Surgery: nasal sinuses | - | 163 | 4 (2.5%) | - |
| Surgery: nasal septum | - | 163 | 8 (4.9%) | - |
| Surgery: nasal turbinates | - | 163 | 1 (0.6%) | - |
| Frequent nasal infections | - | 163 | 20 (12.3%) | - |
| Nasal polyposis | - | 163 | 8 (4.9%) | - |
| Nasal obstruction | - | 163 | 13 (8%) | - |
| Increased nasal secretion | - | 163 | 12 (7.4%) | - |
| Chronic sinusitis | - | 163 | 15 (9.2%) | - |
| Allergic rhinitis | - | 163 | 23 (14.1%) | - |
| Exposure to Toxic Substances | ||||
| Alcohol use | - | 163 | 0 = 23 (14.1%) 1 = 122 (74.8%) 2 = 18 (11%) | - |
| Smoking behavior | - | 163 | 0 = 103 (63.2%) 1 = 37 (22.7%) 2 = 23 (14.1%) | - |
| Professional exposure to chemicals | - | 163 | 23 (14.1%) | - |
| Metabolomics Data | ||||
| Triglycerides in serum | mmol/L | 163 | 0.49–7.9 | 1.57 ± 1.08 |
| Total cholesterol | mmol/L | 163 | 2.8–8.85 | 5.53 ± 1 |
| LDL—low density lipoprotein | mmol/L | 163 | 0.87–6.46 | 3.33 ± 0.83 |
| HDL—high density lipoprotein | mmol/L | 163 | 0.67–2.84 | 1.51 ± 0.43 |
| Hba1c—glycated hemoglobin | mmol/L | 163 | 4.7–6.8 | 5.6 ± 0.4 |
| Glucose, baseline | mmol/L | 163 | 3.88–7.45 | 5.33 ± 0.69 |
| Glucose, after 120 min | mmol/L | 162 | 2.65–16.48 | 6.8 ± 2.31 |
| Glucose, change | mmol/L | 162 | −3.68–9.56 | 1.46 ± 1.99 |
| Fatty acids, baseline | mmol/L | 163 | 0.12–93 | 1.03 ± 7.25 |
| Fatty acids, after 120 min | mmol/L | 162 | 0.01–0.25 | 0.06 ± 0.04 |
| Fatty acids change | mmol/L | 162 | −92.97–0.07 | −0.97 ± 7.28 |
| Proinsulin, baseline | mmol/L | 163 | 0.6–57.2 | 11.31 ± 10.72 |
| Proinsulin, after 120 min | pmol/L | 162 | 0.6–261 | 46.02 ± 50.07 |
| Proinsulin, change | pmol/L | 162 | −0.3–213.7 | 34.66 ± 43.79 |
| C-Peptide, baseline | pmol/L | 163 | 296–3510 | 878.02 ± 390.38 |
| C-Peptide after 120 min | pmol/L | 162 | 500–8165 | 3034.72 ± 1365.23 |
| C-Peptide, change | pmol/L | 162 | −318–6987 | 2155.36 ± 1180.24 |
| Insulin, baseline | pmol/L | 163 | 9–1157 | 79.24 ± 105.04 |
| Insulin, after 120 min | pmol/L | 161 | 26–2455 | 500.27 ± 452.14 |
| Insulin, change | pmol/L | 161 | −293–2271 | 420.97 ± 421.16 |
| Parameter | Classifier Performance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Data | Metabolomics Data Only | |||||||||
| Original | Permuted | |||||||||
| Algorithm | RF | ADA | C5.0 | Regression | SVM | RF | ADA | C5.0 | Regression | SVM |
| Sensitivity, recall | 69 (48.3–86.2) | 65.5 (44.8–82.8) | 65.5 (37.9–86.3) | 72.4 (55.2–89.7) | 72.4 (55.2–86.2) | 58.6 (34.5–79.3) | 51.7 (31–72.4) | 75.9 (13.8–100) | 55.2 (34.4–79.3) | 58.6 (34.5–82.8) |
| Specificity | 64 (44–80) | 60 (40–80) | 64 (28–92) | 64 (44–84) | 64 (44–84) | 40 (16–64) | 48 (24–72) | 24 (0–88) | 42 (16–68) | 40 (16–72) |
| Positive predictive value, precision | 67.7 (57.6–79.2) | 65.8 (53.8–80) | 66.7 (53.8–83.3) | 70 (59.4–82.8) | 70 (59–83.3) | 53.1 (40–68.2) | 53.6 (37.9–69) | 53.7 (33.3–73.3) | 53.3 (36–71.1) | 53.1 (36.4–70.8) |
| Negative predictive value | 62.5 (50–76.9) | 60 (46.4–75) | 60 (46.7–76.2) | 66.7 (54.2–81.3) | 65.5 (53.6–80) | 45.5 (25–68) | 46.2 (28–63.6) | 45.8 (9.5–93.5) | 45.8 (21.7–69.6) | 45.5 (22.2–69.2) |
| F1 | 67.8 (54.9–78) | 65.5 (52–77.2) | 65.5 (47.3–76.9) | 71.4 (60–81.4) | 70.4 (59.3–80) | 56.7 (38.1–71.4) | 53.6 (35.3–68.9) | 64.1 (19–72) | 55.2 (35.1–72.7) | 55.6 (35.1–72.1) |
| Balanced Accuracy | 65 (54.4–75.7) | 63 (50.1–74.5) | 63.3 (50.2–73.9) | 68.2 (56.8–79.4) | 67.9 (56.5–77.9) | 49.3 (34.1–66.2) | 49.9 (33.2–66.5) | 50 (34.1–64.1) | 49.6 (30.7–68.8) | 49.3 (30.7–69) |
| ROC-AUC | 73.3 (62.1–83.2) | 68.8 (53.8–81.7) | 65.2 (51.7–76.7) | 76.4 (64.4–86.8) | 67.9 (56.5–77.9) | 49.2 (28.3–71) | 55.6 (45.5–71.7) | 50 (33.5–65.1) | 60.1 (47–76.8) | 55.3 (37.9–71.1) |
| Data | Metabolomics and Olfactory Data | Olfactory Data Only | ||||||||
| Original | Original | |||||||||
| Sensitivity, recall | 69 (48.3–82.8) | 65.5 (44.8–79.3) | 65.5 (37.9–86.2) | 69 (51.7–86.2) | 69 (51.7–86.2) | 58.6 (41.4–75.9) | 55.2 (37.9–72.4) | 100 (17.2–100) | 69 (44.8–93.1) | 82.8 (41.4–100) |
| Specificity | 64 (44–80) | 64 (40–80) | 60 (36–88) | 64 (44–84) | 64 (44–84) | 44 (24–64) | 48 (28–68) | 0 (0–76) | 28 (8–48) | 16 (0–48) |
| Positive predictive value, precision | 68 (58.1–80) | 66.7 (54.3–78.6) | 65.7 (53.8–81.5) | 69 (59–82.1) | 69 (58.1–81.8) | 55.6 (45.8–65.4) | 55.9 (45–66.7) | 53.7 (41.7–59.1) | 52.6 (44.1–58.5) | 53.5 (44.8–56.3) |
| Negative predictive value | 62.5 (51.5–76.9) | 60 (47.4–73.9) | 60 (46.4–73.9) | 64.3 (52–78.9) | 64 (52.2–78.9) | 50 (35.3–61.5) | 48.3 (35–61.5) | 45.8 (30.8–50) | 43.8 (25–65) | 41.7 (16–62.5) |
| F1 | 67.8 (54.9–78.1) | 65.5 (51.7–76.7) | 65.4 (47.6–75.9) | 69.1 (57.1–80) | 69 (56.6–79.3) | 57.1 (43.6–67.7) | 55.7 (42.3–66.7) | 69.9 (24.4–69.9) | 59.7 (45.2–69.9) | 64.9 (44–69.9) |
| Balanced Accuracy | 65 (54.8–75.1) | 63 (50.8–74.5) | 62.8 (50.1–73.7) | 66.8 (55.6–77.7) | 66.5 (54.8–76.8) | 52.2 (41.3–62.5) | 52.1 (40.4–63.6) | 50 (41.6–54.4) | 48.5 (37.6–57.1) | 49.7 (38.8–53.9) |
| ROC-AUC | 74 (62.8–83.7) | 68.8 (55–81.5) | 65 (52.1–76.1) | 74.5 (63.3–85.5) | 66.5 (54.8–76.8) | 55 (43.3–65.5) | 54.8 (45.4–65.2) | 50 (41.5–54.4) | 54.2 (45.2–64.3) | 50 (40.5–57.3) |
| Parameter |
Classifier Performance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Data | Metabolomics Data Only | Metabolomics and Olfactory Data | ||||||||
| Original | Original | |||||||||
| Algorithm | RF | ADA | C5.0 | Regression | SVM | RF | ADA | C5.0 | Regression | SVM |
| Sensitivity, recall | 0 (0–7.7) | 23.1 (0–46.2) | 0 (0–31) | 0 (0–15.4) | 0 (0–0) | 76.9 (53.8–100) | 76.9 (46.2–92.3) | 76.9 (46.2–100) | 84.6 (53.8–100) | 84.6 (53.8–100) |
| Specificity | 97.6 (90.2–100) | 73.2 (56.1–85.4) | 100 (65.9–100) | 90.2 (75.6–100) | 100 (100–100) | 95.1 (87.8–100) | 92.7 (82.9–100) | 92.7 (80.5–100) | 92.7 (80.5–100) | 95.1 (87.8–100) |
| Positive predictive value, precision | 0 (0–100) | 20 (0–38.5) | 20 (0–40.1) | 8.3 (0–50) | 0 (0–0) | 84.6 (69.2–100) | 78.6 (57.1–100) | 76.9 (55.6–100) | 78.6 (57.1–100) | 85.7 (66.7–100) |
| Negative predictive value | 75.9 (74.5–77.4) | 74.4 (68.4–80.5) | 75.9 (72.1–78) | 75 (71.4–77.8) | 75.9 (75.9–75.9) | 93 (86.9–100) | 92.7 (85.4–97.6) | 92.5 (84.1–100) | 94.9 (86.7–100) | 95 (86.7–100) |
| F1 | 13.3 (11.1–14.3) | 21.4 (7.4–38.7) | 19 (8.3–37.4) | 11.8 (8.3–25) | #WERT! | 81.5 (63.6–92.9) | 76.6 (58.3–88.9) | 76 (52.6–88.9) | 80 (60.8–92.9) | 83.3 (63.6–96.3) |
| Balanced Accuracy | 50 (46.3–53.8) | 46.9 (35.4–58.6) | 50 (41.7–54.4) | 47.6 (40.2–54.2) | 50 (50–50) | 87.2 (74.7–96.3) | 84.3 (71.9–93.7) | 83.6 (68.4–93.7) | 87.4 (73.3–97.6) | 88.6 (74.5–98.8) |
| ROC-AUC | 42.1 (28.1–55.9) | 57 (45.6–72.1) | 50 (40.2–53.4) | 57.6 (45.2–71.1) | 50 (50–50) | 97 (91.7–99.8) | 94.4 (83.3–98.9) | 85.5 (70.6–95.1) | 90.9 (74.9–98.9) | 88.6 (74.5–98.8) |
| Data | Olfactory Data Only | |||||||||
| Original | Permuted | |||||||||
| Sensitivity, recall | 84.6 (53.8–100) | 76.9 (46.2–100) | 76.9 (46.2–100) | 92.3 (69.2–100) | 92.3 (61.5–100) | 7.7 (0–38.5) | 23.1 (0–61.5) | 0 (0–30.8) | 0 (0–15.6) | 0 (0–0) |
| Specificity | 97.6 (90.2–100) | 95.1 (85.4–100) | 95.1 (82.9–100) | 97.6 (92.6–100) | 97.6 (92.7–100) | 92.7 (78–100) | 73.2 (56.1–87.8) | 100 (87.7–100) | 100 (95.1–100) | 100 (100–100) |
| Positive predictive value, precision | 90 (70.6–100) | 81.8 (63.6–100) | 80 (58.8–100) | 91.7 (75–100) | 92.3 (78.6–100) | 22.2 (0–100) | 25 (0–50) | 50 (0–100) | 50 (0–100) | #WERT! |
| Negative predictive value | 95.1 (87–100) | 92.9 (85.4–100) | 92.9 (84.8–100) | 97.5 (90.5–100) | 97.4 (89.1–100) | 75.9 (71.7–82.5) | 76.2 (67.6–85.7) | 75.9 (74.5–80.9) | 75.9 (75.5–78.9) | 75.9 (75.9–75.9) |
| F1 | 84.6 (66.7–96.3) | 78.4 (60–92.3) | 78.6 (55.6–92.3) | 88.9 (74.1–100) | 91.7 (76.2–100) | 20 (9.1–50) | 26.1 (7.1–51.9) | 36.4 (8.3–80) | 14.3 (13.2–63.2) | #WERT! |
| Balanced Accuracy | 89.9 (75.7–98.8) | 85 (72–95.1) | 86 (69.8–96.2) | 93.7 (81–100) | 92.5 (80.8–100) | 50 (40.2–65.4) | 50.6 (34.1–68.6) | 50 (46.3–61.8) | 50 (48.8–57.7) | 50 (50–50) |
| ROC-AUC | 97.8 (93.4–100) | 96.1 (86.1–99.4) | 87 (70.6–96.2) | 98.4 (94.6–100) | 92.5 (80.8–100) | 54.2 (20.3–83.1) | 58.5 (44.5–76.5) | 50 (46.3–64.2) | 81 (48.4–99.4) | 50 (50–50) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lötsch, J.; Hähner, A.; Schwarz, P.E.H.; Tselmin, S.; Hummel, T. Machine Learning Refutes Loss of Smell as a Risk Indicator of Diabetes Mellitus. J. Clin. Med. 2021, 10, 4971. https://doi.org/10.3390/jcm10214971
Lötsch J, Hähner A, Schwarz PEH, Tselmin S, Hummel T. Machine Learning Refutes Loss of Smell as a Risk Indicator of Diabetes Mellitus. Journal of Clinical Medicine. 2021; 10(21):4971. https://doi.org/10.3390/jcm10214971
Chicago/Turabian StyleLötsch, Jörn, Antje Hähner, Peter E. H. Schwarz, Sergey Tselmin, and Thomas Hummel. 2021. "Machine Learning Refutes Loss of Smell as a Risk Indicator of Diabetes Mellitus" Journal of Clinical Medicine 10, no. 21: 4971. https://doi.org/10.3390/jcm10214971
APA StyleLötsch, J., Hähner, A., Schwarz, P. E. H., Tselmin, S., & Hummel, T. (2021). Machine Learning Refutes Loss of Smell as a Risk Indicator of Diabetes Mellitus. Journal of Clinical Medicine, 10(21), 4971. https://doi.org/10.3390/jcm10214971








