Comprehensive Genetic Testing of CYP21A2: A Retrospective Analysis in Patients with Suspected Congenital Adrenal Hyperplasia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Ethical Aspects
2.3. Biochemical Parameters
2.4. Genetic Screening
2.5. Characterization of Variants and Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Study Cohort Characteristics
3.2. Genetic Results
3.3. Genetic Studies of Relatives
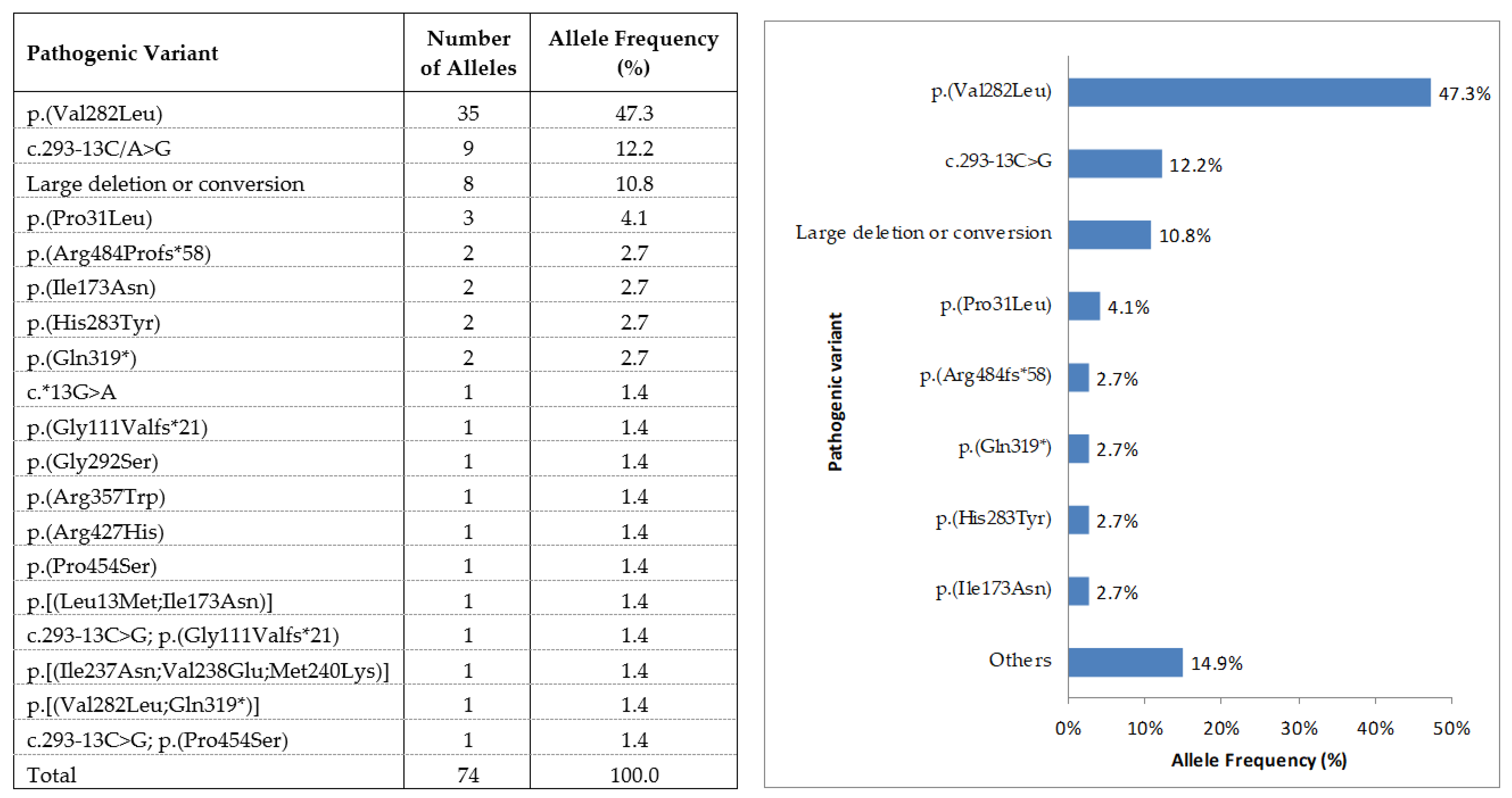
3.4. Allelic Distribution of the Point Mutation Pathological Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carvalho, B.; Marques, C.; Santos-Silva, R.; Fontoura, M.; Carvalho, D.; Carvalho, F. Congenital Adrenal Hyperplasia Due to 21-Hydroxylase Deficiency: An Update on Genetic Analysis of CYP21A2 Gene. Exp. Clin. Endocrinol. Diabetes 2020. [Google Scholar] [CrossRef]
- New, M.I.; Gertner, J.M.; Speiser, P.W.; Del Balzo, P. Growth and final height in classical and nonclassical 21-hydroxylase deficiency. J. Endocrinol. Investig. 1989, 12, 91–95. [Google Scholar]
- Nordenström, A.; Falhammar, H. MANAGEMENT OF ENDOCRINE DISEASE: Diagnosis and management of the patient with non-classic CAH due to 21-hydroxylase deficiency. Eur. J. Endocrinol. 2019, 180, R127–R145. [Google Scholar] [CrossRef]
- New, M.I.; Abraham, M.; Gonzalez, B.; Dumic, M.; Razzaghy-Azar, M.; Chitayat, D.; Sun, L.; Zaidi, M.; Wilson, R.C.; Yuen, T. Genotype-phenotype correlation in 1,507 families with congenital adrenal hyperplasia owing to 21-hydroxylase deficiency. Proc. Natl. Acad. Sci. USA 2013, 110, 2611–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haider, S.; Islam, B.; D’Atri, V.; Sgobba, M.; Poojari, C.; Sun, L.; Yuen, T.; Zaidi, M.; New, M.I. Structure-phenotype correlations of human CYP21A2 mutations in congenital adrenal hyperplasia. Proc. Natl. Acad. Sci. USA 2013, 110, 2605–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merke, D.P.; Bornstein, S.R. Congenital adrenal hyperplasia. Lancet 2005, 365, 2125–2136. [Google Scholar] [CrossRef]
- Parajes, S.; Quinteiro, C.; Domínguez, F.; Loidi, L. High frequency of copy number variations and sequence variants at CYP21A2 locus: Implication for the genetic diagnosis of 21-hydroxylase deficiency. PLoS ONE 2008, 3, e2138. [Google Scholar] [CrossRef] [PubMed]
- White, P.C.; New, M.I.; Dupont, B. Adrenal 21-hydroxylase cytochrome P-450 genes within the MHC class III region. Immunol. Rev. 1985, 87, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Ezquieta, B.; Oliver, A.; Gracia, R.; Gancedo, P.G. Analysis of steroid 21-hydroxylase gene mutations in the Spanish population. Hum. Genet. 1995, 96, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Loidi, L.; Quinteiro, C.; Parajes, S.; Barreiro, J.; Lestón, D.G.; Cabezas-Agricola, J.M.; Sueiro, A.M.; Araújo-Vilar, D.; Catro-Feijoo, L.; Costas, J.; et al. High variability in CYP21A2 mutated alleles in Spanish 21-hydroxylase deficiency patients, six novel mutations and a founder effect. Clin. Endocrinol. 2006, 64, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Lobato, M.N.; Aledo, R.; Meseguer, A. High variability of CYP21 gene rearrangements in Spanish patients with classic form of congenital adrenal hyperplasia. Hum. Hered. 1998, 48, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Morreale, H.F.; Sanchon, R.; Millan, J.L.S. A prospective study of the prevalence of nonclassical congenital adrenal hyperplasia among women presenting with hyperandrogenic symptoms and signs. J. Clin. Endocrinol. Metab. 2008, 93, 527–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcantara-Aragon, V.; Martinez-Couselo, S.; Tundidor-Rengel, D.; Webb, S.M.; Carreras, G.; Espinos, J.M.; Chico, A.; Blanco-Vaca, F.; Corcoy, R. Genetic analysis does not confirm non-classical congenital adrenal hyperplasia in more than a third of the women followed with this diagnosis. Hormones 2014, 13, 585–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speiser, P.W.; Arlt, W.; Auchus, R.J.; Baskin, L.S.; Conway, G.S.; Merke, D.P.; Meyer-Bahlburg, H.F.L.; Miller, W.L.; Murad, M.H.; Oberfield, S.E.; et al. Congenital Adrenal Hyperplasia Due to Steroid 21-Hydroxylase Deficiency: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2018, 103, 4043–4088. [Google Scholar] [CrossRef] [PubMed]
- Votava, F.; Novotna, D.; Kracmar, P.; Vinohradska, H.; Stahlova-Hrabincova, E.; Vrzalová, Z.; Neumann, D.; Malikova, J.; Lebl, J.; Matern, D. Lessons learned from 5 years of newborn screening for congenital adrenal hyperplasia in the Czech Republic: 17-hydroxyprogesterone, genotypes, and screening performance. Eur. J. Pediatr. 2012, 171, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Staden, R.; Beal, K.; Bonfield, J. The Staden package, 1998. Methods Mol. Biol. 2000, 132, 115–130. [Google Scholar] [PubMed]
- Baumgartner-Parzer, S.; Witsch-Baumgartner, M.; Hoeppner, W. EMQN best practice guidelines for molecular genetic testing and reporting of 21-hydroxylase deficiency. Eur. J. Hum. Genet. 2020, 28, 1341–1367. [Google Scholar] [CrossRef]
- Tusie-Luna, M.T.; Traktman, P.; White, P.C. Determination of functional effects of mutations in the steroid 21-hydroxylase gene (CYP21) using recombinant vaccinia virus. J. Biol. Chem. 1990, 265, 20916–20922. [Google Scholar] [CrossRef]
- Higashi, Y.; Hiromasa, T.; Tanae, A.; Miki, T.; Nakura, J.; Kondo, T.; Ohura, T.; Ogawa, E.; Nakayama, K.; Fujii-Kuriyama, Y. Effects of individual mutations in the P-450(C21) pseudogene on the P-450(C21) activity and their distribution in the patient genomes of congenital steroid 21-hydroxylase deficiency. J. Biochem. 1991, 109, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Nikoshkov, A.; Lajic, S.; Vlamis-Gardikas, A.; Tranebjærg, L.; Holst, M.; Wedell, A.; Luthman, H. Naturally occurring mutants of human steroid 21-hydroxylase (P450c21) pinpoint residues important for enzyme activity and stability. J. Biol. Chem. 1998, 273, 6163–6165. [Google Scholar] [CrossRef] [Green Version]
- Neocleous, V.; Fanis, P.; Toumba, M.; Stylianou, C.; Picolos, M.; Andreou, E.; Kyriakou, A.; Iasonides, M.; Nicolaou, S.; Kyriakides, T.C.; et al. The Spectrum of Genetic Defects in Congenital Adrenal Hyperplasia in the Population of Cyprus: A Retrospective Analysis. Horm. Metab. Res. 2019, 51, 586–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosse, S.D.; Van Vliet, G. How many deaths can be prevented by newborn screening for congenital adrenal hyperplasia? Horm. Res. 2007, 67, 284–291. [Google Scholar] [CrossRef]
- Speiser, P.W.; White, P.C. Congenital adrenal hyperplasia. N. Engl. J. Med. 2003, 349, 776–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, P.C.; Speiser, P.W. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr. Rev. 2000, 21, 245–291. [Google Scholar] [CrossRef]
- Forest, M.G. Recent advances in the diagnosis and management of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Hum. Reprod. Update 2004, 10, 469–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Concolino, P.; Costella, A. Congenital Adrenal Hyperplasia (CAH) due to 21-Hydroxylase Deficiency: A Comprehensive Focus on 233 Pathogenic Variants of CYP21A2 Gene. Mol. Diagn. Ther. 2018, 22, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Finkielstain, G.P.; Chen, W.; Mehta, S.P.; Fujimura, F.K.; Hanna, R.M.; Van Ryzin, C.; McDonnell, N.B.; Merke, D.P. Comprehensive genetic analysis of 182 unrelated families with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J. Clin. Endocrinol. Metab. 2011, 96, E161–E172. [Google Scholar] [CrossRef] [PubMed]
- Sarafoglou, K.; Lorentz, C.P.; Otten, N.; Oetting, W.S.; Grebe, S.K.G. Molecular testing in congenital adrenal hyperplasia due to 21alpha-hydroxylase deficiency in the era of newborn screening. Clin. Genet. 2012, 82, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Krone, N.; Braun, A.; Roscher, A.A.; Knorr, D.; Schwarz, H.P. Predicting phenotype in steroid 21-hydroxylase deficiency? Comprehensive genotyping in 155 unrelated, well defined patients from southern Germany. J. Clin. Endocrinol. Metab. 2000, 85, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Dracopoulou-Vabouli, M.; Maniati-Christidi, M.; Dacou-Voutetakis, C. The spectrum of molecular defects of the CYP21 gene in the Hellenic population: Variable concordance between genotype and phenotype in the different forms of congenital adrenal hyperplasia. J. Clin. Endocrinol. Metab. 2001, 86, 2845–2848. [Google Scholar] [PubMed] [Green Version]
- Sadeghi, F.; Yurur-Kutlay, N.; Berberoglu, M.; Cetinkaya, E.; Aycan, Z.; Kara, C.; Ruhi, H.I.; Ocal, G.; Şıklar, Z.; Elhan, A.; et al. Identification of frequency and distribution of the nine most frequent mutations among patients with 21-hydroxylase deficiency in Turkey. J. Pediatr. Endocrinol. Metab. 2008, 21, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Carrera, P.; Bordone, L.; Azzani, T.; Brunelli, V.; Garancini, M.P.; Chiumello, G.; Ferrari, M. Point mutations in Italian patients with classic, non-classic, and cryptic forms of steroid 21-hydroxylase deficiency. Hum. Genet. 1996, 98, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L. MECHANISMS IN ENDOCRINOLOGY: Rare defects in adrenal steroidogenesis. Eur. J. Endocrinol. 2018, 179, R125–R141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witchel, S.F. Non-classic congenital adrenal hyperplasia. Steroids 2013, 78, 747–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadakis, G.; Kandaraki, E.A.; Tseniklidi, E.; Papalou, O.; Diamanti-Kandarakis, E. Polycystic Ovary Syndrome and NC-CAH: Distinct Characteristics and Common Findings. A Systematic Review. Front. Endocrinol. 2019, 10, 388. [Google Scholar] [CrossRef] [PubMed]
- Kleinle, S.; Lang, R.; Fischer, G.F.; Vierhapper, H.; Waldhauser, F.; Fodinger, M.; Baumgartner-Parzer, S.M. Duplications of the functional CYP21A2 gene are primarily restricted to Q318X alleles: Evidence for a founder effect. J. Clin. Endocrinol. Metab. 2009, 94, 3954–3958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azziz, R.; Dewailly, D.; Owerbach, D. Clinical review 56: Nonclassic adrenal hyperplasia: Current concepts. J. Clin. Endocrinol. Metab. 1994, 78, 810–815. [Google Scholar] [PubMed]
- Concolino, P. Issues with the Detection of Large Genomic Rearrangements in Molecular Diagnosis of 21-Hydroxylase Deficiency. Mol. Diagn. Ther. 2019, 23, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Merke, D.P.; Auchus, R.J. Congenital Adrenal Hyperplasia Due to 21-Hydroxylase Deficiency. N. Engl. J. Med. 2020, 383, 1248–1261. [Google Scholar] [CrossRef] [PubMed]
| Classic SW | Classic SV | NC | Non-Definite | Negative Genetic Diagnosis | |
|---|---|---|---|---|---|
| Patients n (%) | 5 (8.5) | 2 (3.4) | 29 (49.2) | 1 (1.7) | 22 (37.9) |
| Age at diagnosis in years, mean (SD) | 0.043 (0.035) | 16.5 (21.9) | 21.3 (10.9) | 3 | 15.7 (9.1) |
| Female (%) | 3 (60) | 2 (100) | 27 (93.1) | 0 (0) | 20 (91) |
| Clinical features at presentation, n (%) | |||||
| Adrenal crisis | 5 (100.0) | 0 | - | - | - |
| Atypical genitalia | 3 (60.0) | 2 (100.0) | - | - | 2 (9.1) |
| Early puberty | - | - | 6 (20.7) | 1 (100) | 3 (13.6) |
| Hirsutism | - | - | 7 (24.1) | - | 4 (18.2) |
| Acne | - | - | 1 (3.4) | - | 1 (4.5) |
| Menstrual abnormalities | - | - | 3 (10.3) | - | 4 (18.2) |
| Infertility | - | - | 7 (24.1) | - | 2 (9.1) |
| 17-OHP concentration (nmol/L) | |||||
| 17-OHP (basal), median (IQR) | 60.6 (60.6–60.6) | 60.6 (60.6–60.6) | 41.2 (19.8–59.2) | 60.6 | 6.2 (3.1–8.8) |
| 17-OHP (60 min post ACTH), median (IQR) | - | - | 60.6 (60.6–60.6) | - | 16.3 (7.7–19.8) |
| Mutation status n (%) | |||||
| Homozygosis | - | - | 10 (34.5) | - | - |
| Compound Heterozygosis | 2 (40.0) | 2 (100.0) | 16 (55.2) | 1 (100.0) | - |
| Hemizygosis | 3 (60.0) | - | 3 (10.3) | - | - |
| Heterozygote carriers | - | - | - | - | 11 (50.0) |
| Wild type | - | - | - | - | 11 (50.0) |
| Group | Enzyme Activity | Pathogenic Variants (DNA) | Pathogenic Variants (Protein) | Proband | Number of Patients | Expected Phenotype | Observed Phenotype |
|---|---|---|---|---|---|---|---|
| 0 | Null | c.[(-1)_(202+1_203-1)del];[(-1)_(202+1_203-1)del;(651+1_652-1)_(313+1_314-1)del] | p.[?];[?] | Compound Hz | 1 | SW | SV |
| A | <2% | c.[293-13C>G];[(-1)-(1118+1_119-1)del] | p.[?];[?] | Hemizygote | 2 | SW | SW |
| c.[1069C>T];[(-1)-(1118+1_119-1)del] | p.[(Arg357Trp)];[?] | Hemizygote | 1 | SW | SW | ||
| c.[293-13C>G];[874G>A] | p.[?];[(Gly292Ser)] | Compound Hz | 1 | SW | SW | ||
| c.[293-13C>G];[710T>A;713T>A;719T>A] | p.[?];[(Ile237Asn;Val238Glu;Met240Lys)] | Compound Hz | 1 | SW | SV | ||
| B | ~2% | c.[293-13C>G];[518T>A] | p.[?];[(Ile173Asn)] | Compound Hz | 1 | SV | NC/SV |
| c.[37C>A;518T>A];[955C>T] | p.[(Leu13Met;p.Ile173Asn];[(Gln319*)] | Compound Hz | 1 | SV | SW | ||
| C | ~20–60% | c.[844G>T];[844G>T] | p.[(Val282Leu)];[(Val282Leu)] | Hm | 9 | NC | NC |
| c.[844G>T];[(-1)-(1118+1_119-1)del] | p.[(Val282Leu)];[?] | Hemizygote | 3 | NC | NC | ||
| c.[844G>T];[955C>T] | p.[(Val282Leu)];[(Gln319*)] | Compound Hz | 1 | NC | NC | ||
| c.[844G>T];[1280G>A] | p.[(Val282Leu)];[(Arg427His)] | Compound Hz | 1 | NC | NC | ||
| c.[844G>T];[1360C>T] | p.[(Val282Leu)];[(Pro454Ser)] | Compound Hz | 1 | NC | NC | ||
| c.[844G>T];[1451_1452delinsC] | p.[(Val282Leu)];[(Arg484Profs*58)] | Compound Hz | 1 | NC | NC | ||
| c.[844G>T];[ 955C>T; 1451_1452delinsC] | p.[(Val282Leu;Gln319*];[(Arg484Profs*58)] | Compound Hz | 1 | NC | NC | ||
| c.[92C>T];[844G>T] | p.[(Pro31Leu)];[(Val282Leu)] | Compound Hz | 2 | NC | NC | ||
| c.[92C>T];[518T>A] | p.[(Pro31Leu)];[(Ile173Asn)] | Compound Hz | 1 | NC | NC | ||
| c.[332_339del];[844G>T] | p.[(Gly111Valfs*21)];[(Val282Leu)] | Compound Hz | 1 | NC | NC | ||
| c.[293-13C>G];[844G>T] | p.[?];[(Val282Leu)] | Compound Hz | 1 | NC | NC | ||
| c.[293-13C>G];[844G>T] | p.[?];[(Val282Leu)] | Compound Hz | 3 | NC | NC | ||
| c.[293-13C>G;332-339del];[844G>T] | p.[(?; Gly111Valfs*21)];[(Val282Leu)] | Compound Hz | 1 | NC | NC | ||
| c.[293-13C>G;1360C>T];[844G>T] | p.[(?; Pro454Ser)];[(Val282Leu)] | Compound Hz | 1 | NC | NC | ||
| c.[844G>T];[*13G>A] | p.[?];[(Val282Leu)] | Compound Hz | 1 | NC | NC | ||
| D | Unknown | c.[847C>T];[847C>T] | p.[(His283Tyr)];[(His283Tyr)] | Hm | 1 | - | NC |
| Patient | Cis/Trans | Genotype | Phenotype |
|---|---|---|---|
| 1 | trans | c.[293-13C>G];p.[(Ile237Asn;Val238Glu;Met240Lys)] | SV |
| 2 | trans | c.[293-13C>G];p.[(Val282Leu)] | NC |
| 3 | trans | c.[293-13C>G];p.[(Val282Leu)] | NC |
| 4 | trans | c.[293-13C>G];p.[(Val282Leu)] | NC |
| 5 | trans | c.[293-13C>G];p.[(Ile173Asn)] | NC |
| 6 | cis | [c.293-13C>G;p.(Pro454Ser)] | NC |
| 7 | trans | p.[(Val282Leu)];[(Pro454Ser)] | NC |
| 8 | trans | p.[(Pro31Leu)];[(Val282Leu)] | NC |
| 9 | trans | p.[(Pro31Leu)];[(Val282Leu)] | NC |
| 10 | trans | p.[(Val282Leu)];[(Arg427His)] | NC |
| 11 | trans | p.[(Gly111Valfs*21)];[(Val282Leu)] | NC |
| 12 | trans | c.[13*G>A];p.[(Val282Leu)] | NC |
| 13 | trans | [c.293-13C>G;p.(Gly111Valfs*21)];[p.(Val282Leu)] | NC |
| 14 | trans | [c.293-13C>G;p.(Pro454Ser)];[p.(Val282Leu)] | NC |
| 15 | trans | p.[(Val282Leu)];[(Gln319*;Arg484Profs*58)] | NC |
| 16 | trans | p.[(Leu13Met;Ile173Asn)];[(Glu319*)] | SW |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nan, M.N.; Roig, R.; Martínez, S.; Rives, J.; Urgell, E.; Espinós, J.J.; Tirado, M.; Carreras, G.; Aulinas, A.; Webb, S.M.; et al. Comprehensive Genetic Testing of CYP21A2: A Retrospective Analysis in Patients with Suspected Congenital Adrenal Hyperplasia. J. Clin. Med. 2021, 10, 1183. https://doi.org/10.3390/jcm10061183
Nan MN, Roig R, Martínez S, Rives J, Urgell E, Espinós JJ, Tirado M, Carreras G, Aulinas A, Webb SM, et al. Comprehensive Genetic Testing of CYP21A2: A Retrospective Analysis in Patients with Suspected Congenital Adrenal Hyperplasia. Journal of Clinical Medicine. 2021; 10(6):1183. https://doi.org/10.3390/jcm10061183
Chicago/Turabian StyleNan, Madalina Nicoleta, Rosa Roig, Susana Martínez, Jose Rives, Eulàlia Urgell, Juan José Espinós, Mireia Tirado, Gemma Carreras, Anna Aulinas, Susan M. Webb, and et al. 2021. "Comprehensive Genetic Testing of CYP21A2: A Retrospective Analysis in Patients with Suspected Congenital Adrenal Hyperplasia" Journal of Clinical Medicine 10, no. 6: 1183. https://doi.org/10.3390/jcm10061183






