Assessment of Liver Fibrosis Stage Using Integrative Analysis of Hepatic Heterogeneity and Nodularity in Routine MRI with FIB-4 Index as Reference Standard
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statment
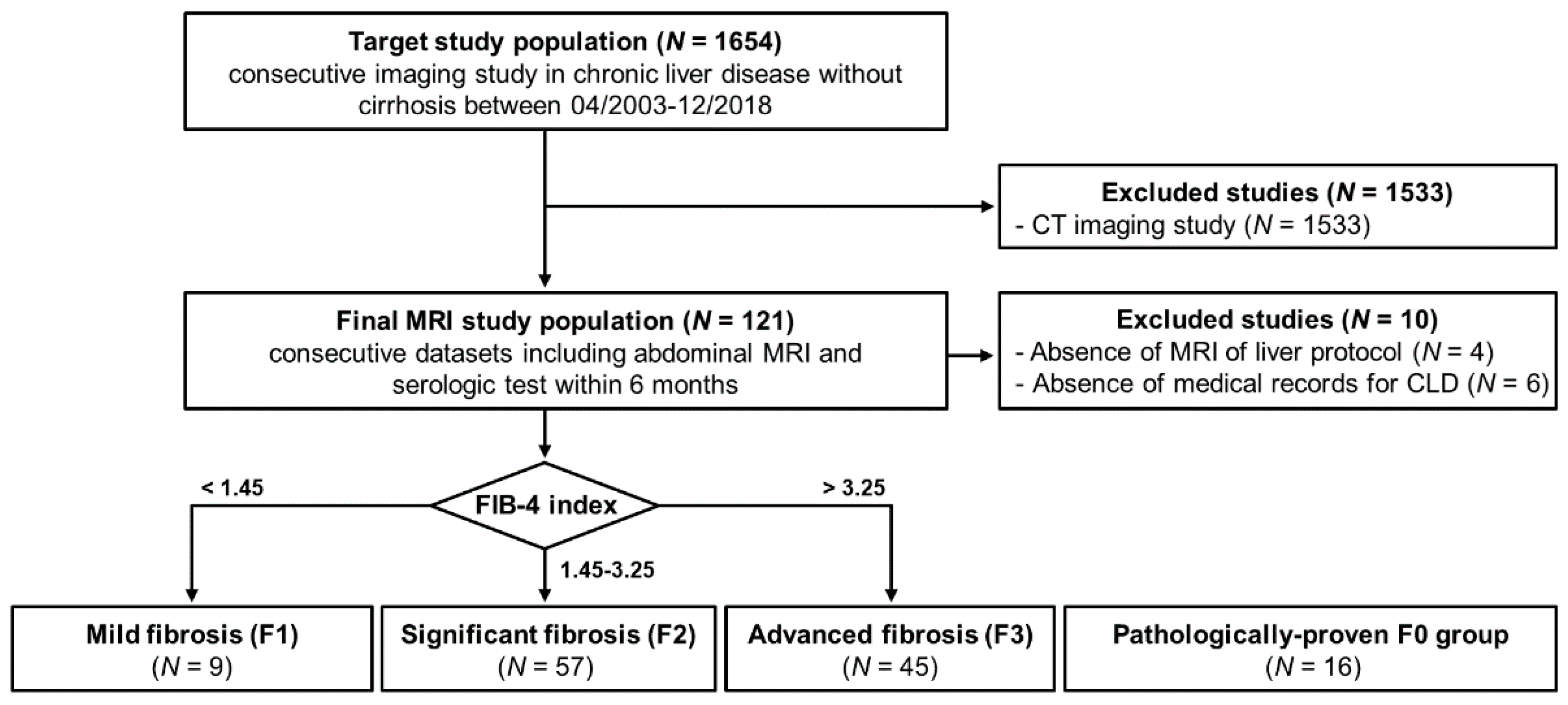
2.2. Subject Population
2.3. Acquisition of MRI
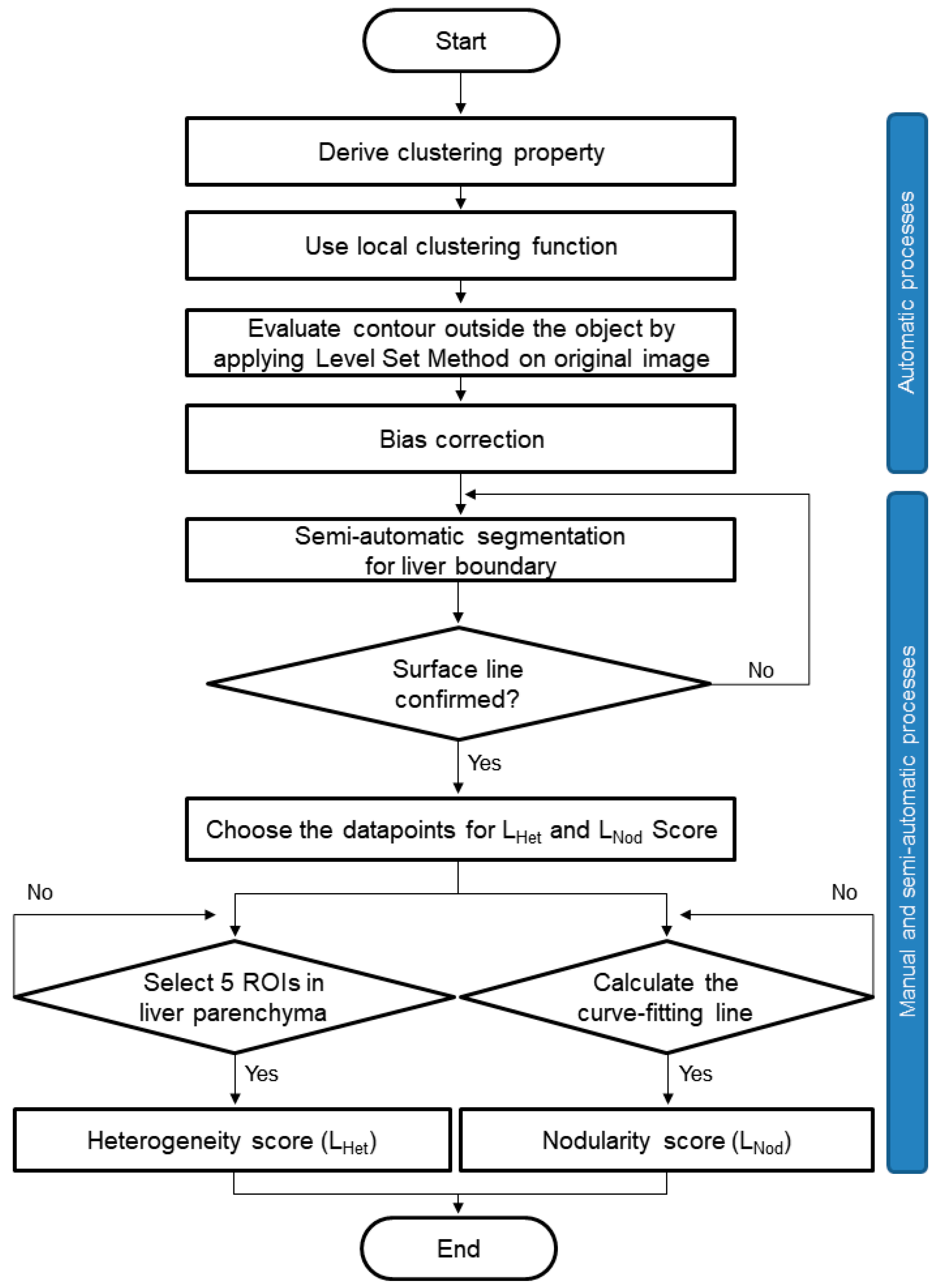
2.4. Software for Quantification of Liver Heterogeneity and Nodularity
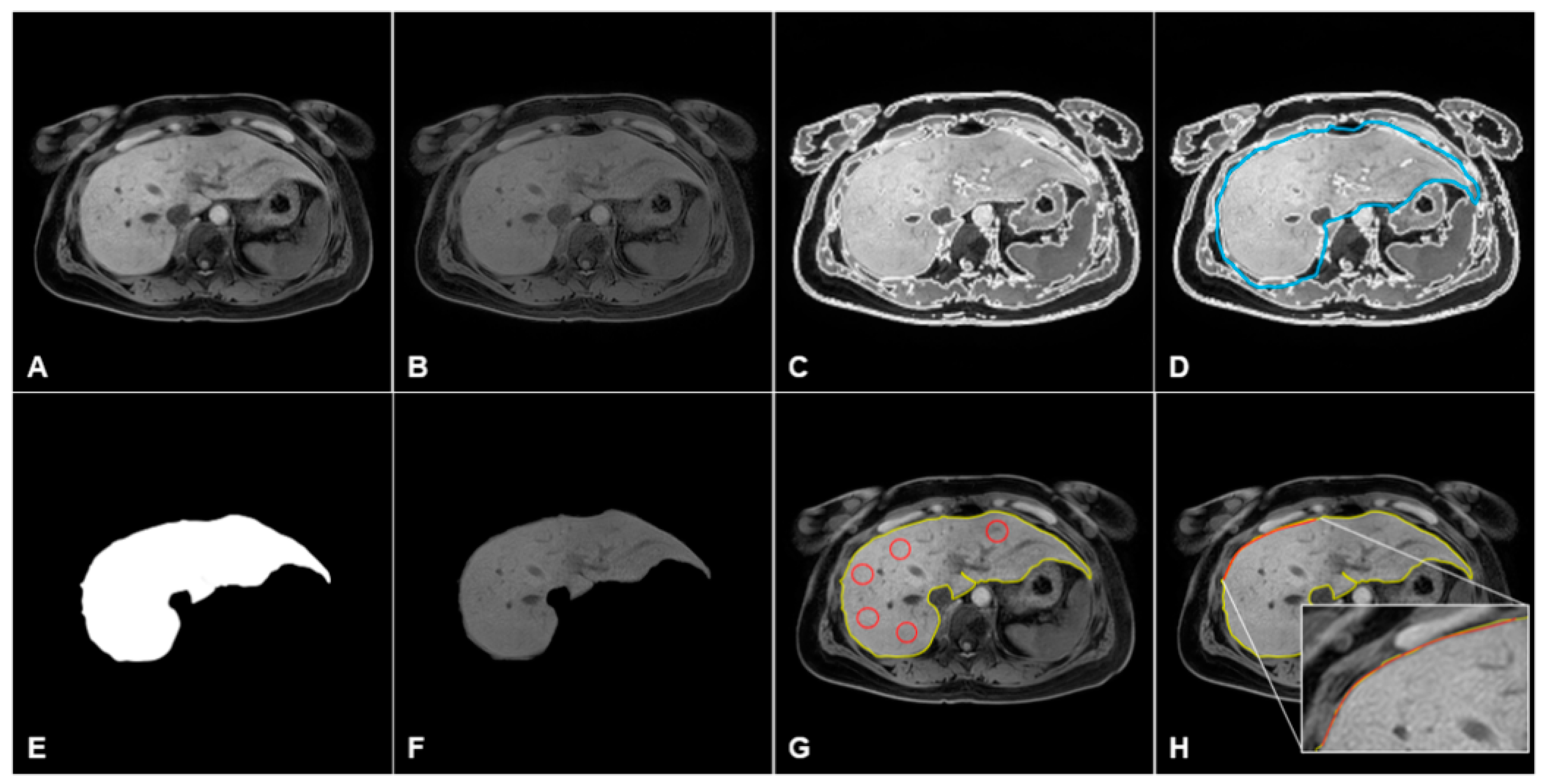
2.5. Data Processing and Quantification of MRI in CLD
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics in Fibrosis Stages
3.2. Liver Heterogeneity and Nodularity Measurements in CLD
3.3. ROC Analysis for Differential Diagnosis According to Fibrosis Stages
3.4. Intraobserver and Interobserver Agreement
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karsdal, M.A.; Manon-Jensen, T.; Genovese, F.; Kristensen, J.H.; Nielsen, M.J.; Sand, J.M.; Hansen, N.U.; Bay-Jensen, A.C.; Bager, C.L.; Krag, A.; et al. Novel insights into the function and dynamics of extracellular matrix in liver fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G807–G830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebel, N.H.; Horslen, S.P. Complications and management of chronic liver disease. In Diseases of the Liver and Biliary System in Children, 4th ed.; Kelly, D.A., Ed.; John Wiley & Sons Ltd.: New York, NY, USA, 2017. [Google Scholar]
- Embade, N.; Millet, O. Molecular Determinants of Chronic Liver Disease as Studied by NMR-Metabolomics. Curr. Top. Med. Chem. 2017, 17, 2752–2766. [Google Scholar] [CrossRef] [PubMed]
- Bedossa, P.; Carrat, F. Liver biopsy: The best, not the gold standard. J. Hepatol. 2009, 50, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, I.Y.; Catalano, O.A.; Caravan, P. Advances in functional and molecular MRI technologies in chronic liver diseases. J. Hepatol. 2020, 73, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Martínez, S.M.; Crespo, G.; Navasa, M.; Forns, X. Noninvasive assessment of liver fibrosis. Hepatology 2011, 53, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Porter, K.K.; Elkassem, A.A.; Sanyal, R.; Lockhart, M.E. Current imaging techniques for noninvasive staging of hepatic fibrosis. AJR Am. J. Roentgenol. 2019, 213, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.P.; Venkatesh, S.K. Imaging of Hepatic Fibrosis. Curr. Gastroenterol. Rep. 2018, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Petitclerc, L.; Sebastiani, G.; Gilbert, G.; Cloutier, G.; Tang, A. Liver fibrosis: Review of current imaging and MRI quantification techniques. J. Magn. Reson. Imaging 2017, 45, 1276–1295. [Google Scholar] [CrossRef]
- Lee, G.M.; Kim, Y.R.; Ryu, J.H.; Kim, T.H.; Cho, E.Y.; Lee, Y.H.; Yoon, K.H. Quantitative measurement of hepatic fibrosis with gadoxetic acid-enhanced magnetic resonance imaging in patients with chronic hepatitis B Infection: A comparative study on aspartate aminotransferase to platelet ratio index and fibrosis-4 index. Korean J. Radiol. 2017, 18, 444–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Kim, Y.R.; Lee, G.M.; Ryu, J.H.; Cho, E.Y.; Lee, Y.H.; Yoon, K.H. Coefficient of variation on Gd-EOB MR imaging: Correlation with the presence of early-stage hepatocellular carcinoma in patients with chronic hepatitis B. Eur. J. Radiol. 2018, 102, 95–101. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, J.E.; Ryu, J.H.; Jeong, C.W. Development of liver surface nodularity quantification program and its clinical application in nonalcoholic fatty liver disease. Sci. Rep. 2019, 9, 9994. [Google Scholar] [CrossRef]
- Smith, A.D.; Branch, C.R.; Zand, K.; Subramony, C.; Zhang, H.; Thaggard, K.; Hosch, R.; Bryan, J.; Vasanji, A.; Griswold, M.; et al. Liver surface nodularity quantification from routine CT images as a biomarker for detection and evaluation of cirrhosis. Radiology 2016, 280, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Pickhardt, P.J.; Malecki, K.; Kloke, J.; Lubner, M.G. Accuracy of liver surface nodularity quantification on MDCT as a noninvasive biomarker for staging hepatic fibrosis. AJR Am. J. Roentgenol. 2016, 207, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Besa, C.; Wagner, M.; Lo, G.; Gordic, S.; Chatterji, M.; Kennedy, P.; Stueck, A.; Thung, S.; Babb, J.; Smith, A.; et al. Detection of liver fibrosis using qualitative and quantitative MR elastography compared to liver surface nodularity measurement, gadoxetic acid uptake, and serum markers. J. Magn. Reson. Imaging 2018, 47, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Zand, K.A.; Florez, E.; Sirous, R.; Shlapak, D.; Souza, F.; Roda, M.; Bryan, J.; Vasanji, A.; Griswold, M.; et al. Liver surface nodularity score allows prediction of cirrhosis decompensation and death. Radiology 2017, 283, 711–722. [Google Scholar] [CrossRef]
- Huang, G.; Ji, H.; Zhang, W. A fast level set method for inhomogeneous image segmentation with adaptive scale parameter. Magn. Reson. Imaging 2018, 52, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Rozario, L.J.; Moazzam, M.G.; Begum, M.; Naowar, F.F. An Improved Method of Segmentation of Medical Images thatc Incorporates Bias Correction. Int. J. Eng. Sci. 2014, 3, 1–8. [Google Scholar]
- Kim, T.H.; Jun, H.Y.; Kim, K.J.; Lee, Y.H.; Lee, M.S.; Choi, K.H.; Yun, K.J.; Jeong, Y.Y.; Jun, C.H.; Cho, E.Y.; et al. Hepatic alanine differentiates nonalcoholic steatohepatitis from simple steatosis in humans and mice: A proton MR spectroscopy study with long echo time. J. Magn. Reson. Imaging 2017, 46, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, J.; Liu, Q.; Li, C.; Liu, Z.; Wang, M.; Zheng, H.; Wang, S. A Radiomics Approach With CNN for Shear-Wave Elastography Breast Tumor Classification. IEEE Trans. Biomed. Eng. 2018, 65, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Yang, J.; Yan, L. Comparison of diagnostic accuracy of aspartate aminotransferase to platelet ratio index and fibrosis-4 index for detecting liver fibrosis in adult patients with chronic hepatitis B virus infection: A systemic review and meta-analysis. Hepatology 2015, 61, 292–302. [Google Scholar] [CrossRef]
- de Oliveira, A.C.; El-Bacha, I.; Vianna, M.V.; Parise, E.R. Utility and limitations of APRI and FIB4 to predict staging in a cohort of nonselected outpatients with hepatitis C. Ann. Hepatol. 2016, 15, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.-H.; Kuo, F.-Y.; Kee, K.-M.; Chang, K.-C.; Tsai, M.-C.; Hu, T.-H.; Lu, S.-N.; Wang, J.-H.; Hung, C.-H.; Chen, C.-H. APRI and FIB-4 in the evaluation of liver fibrosis in chronic hepatitis C patients stratified by AST level. PLoS ONE 2018, 13, e0199760. [Google Scholar] [CrossRef]
- Jang, H.J.; Min, J.H.; Lee, J.E.; Shin, K.S.; Kim, K.H.; Choi, S.Y. Assessment of liver fibrosis with gadoxetic acid-enhanced MRI: Comparisons with transient elastography, ElastPQ, and serologic fibrosis markers. Abdom. Imaging 2019, 44, 2769–2780. [Google Scholar] [CrossRef] [PubMed]
- Feier, D.; Balassy, C.; Bastati, N.; Fragner, R.; Wrba, F.; Ba-Ssalamah, A. The diagnostic efficacy of quantitative liver MR imaging with diffusion-weighted, SWI, and hepato-specific contrast-enhanced sequences in staging liver fibrosis--a multiparametric approach. Eur. Radiol. 2016, 26, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Lee, S.S.; Park, B.; Yun, J.; Sung, Y.S.; Shim, W.H.; Shin, Y.M.; Kim, S.Y.; Lee, S.J.; Lee, M.G. Radiomics analysis of gadoxetic acid-enhanced MRI for staging liver fibrosis. Radiology 2019, 290, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Raunig, D.L.; McShane, L.M.; Pennello, G.; Gatsonis, C.; Carson, P.L.; Voyvodic, J.T.; Wahl, R.L.; Kurland, B.F.; Schwarz, A.J.; Gonen, M.; et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Stat. Methods Med. Res. 2015, 24, 27–67. [Google Scholar] [CrossRef] [Green Version]
- Sartoris, R.; Rautou, P.E.; Elkrief, L.; Pollorsi, G.; Durand, F.; Valla, D.; Spahr, L.; Terraz, S.; Soubrane, O.; Cauchy, F.; et al. Quantification of liver surface nodularity at CT: Utility for detection of portal hypertension. Radiology 2018, 289, 698–707. [Google Scholar] [CrossRef] [PubMed]



| F0 (n = 16) | F1 (n = 9) | F2 (n = 57) | F3 (n = 45) | p-Value * † | Multiple Comparisons | |||
|---|---|---|---|---|---|---|---|---|
| a F1 vs. F2 | b F1 vs. F3 | c F2 vs. F3 | ||||||
| Etiology * | ||||||||
| Unknown | 16 (100) | - | - | - | ||||
| Hepatitis B | - | 6 (67) | 37 (65) | 27 (60) | * 0.570 | |||
| Hepatitis C | - | 0 (0) | 5 (9) | 4 (9) | ||||
| Coinfection | - | 0 (0) | 2 (4) | 0 (0) | ||||
| Alcohol | - | 1 (11) | 5 (9) | 6 (13) | ||||
| NAFLD | - | 0 (0) | 6 (10) | 5 (11) | ||||
| Autoimmune | - | 2 (22) | 2 (3) | 3 (7) | ||||
| Serum biochemistry † | ||||||||
| Age (year) | 35.0 ± 15.5 | 50.3 ± 14.9 | 60.3 ± 12.1 | 64.8 ± 13.6 | † 0.008 b | 0.086 | 0.008 | 0.191 |
| BMI | 24.8 ± 3.2 | 24.8 ± 1.7 | 24.6 ± 2.0 | 25.3 ± 2.9 | † 0.211 | 0.953 | 0.777 | 0.184 |
| Albumin (g/dL) | 4.36 ± 0.07 | 3.80 ± 0.31 | 4.08 ± 0.43 | 4.30 ± 0.69 | † 0.873 | 0.962 | 0.888 | 0.928 |
| ALP (IU/L) | 73.9 ± 8.3 | 279.2 ± 36.7 | 285.0 ± 17.9 | 459.8 ± 53.6 | † 0.002 c | 0.998 | 0.125 | 0.002 |
| ALT (IU/L) | 18.8 ± 2.1 | 31.7 ± 8.7 | 37.5 ± 3.8 | 74.2 ± 25.6 | † 0.224 | 0.989 | 0.552 | 0.230 |
| AST (IU/L) | 21.0 ± 1.4 | 26.0 ± 4.4 | 38.7 ± 3.6 | 109.1 ± 36.0 | † 0.058 | 0.972 | 0.311 | 0.064 |
| Bilirubin (mg/dL) | 0.64 ± 0.09 | 0.73 ± 0.09 | 0.78 ± 0.09 | 2.66 ± 0.90 | † 0.058 | 1.000 | 0.441 | 0.052 |
| GGT (IU/L) | 38.5 ± 10.9 | 83.4 ± 29.9 | 63.2 ± 7.2 | 184.9 ± 30.0 | † 0.001 c | 0.946 | 0.266 | <0.001 |
| Platelet count (103/μL) | 243.5 ± 8.2 | 223.9 ± 17.4 | 172.3 ± 7.0 | 135.4 ± 6.9 | <† 0.001 a b c | 0.014 | <0.001 | 0.001 |
| F0 (n = 16) | F1 (n = 9) | F2 (n = 57) | F3 (n = 45) | p-Value * | Multiple Comparisons | |||
|---|---|---|---|---|---|---|---|---|
| a F1 vs. F2 | b F1 vs. F3 | c F2 vs. F3 | ||||||
| Heterogeneity (LHet, %) | 3.49 ± 0.34 | 5.52 ± 0.88 | 6.74 ± 0.89 | 7.56 ± 1.79 | <0.001a b c | 0.032 | <0.001 | 0.008 |
| Nodularity (LNod) | 0.84 ± 0.06 | 0.91 ± 0.04 | 1.09 ± 0.08 | 1.14 ± 0.14 | <0.001 a b c | <0.001 | <0.001 | 0.022 |
| LHet × LNod score | 2.96 ± 0.46 | 5.01 ± 0.91 | 7.30 ± 0.89 | 8.48 ± 1.34 | <0.001 a b c | <0.001 | <0.001 | <0.001 |
| Comparison | Threshold Value | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | DA (%) | AUROC | p-Value |
|---|---|---|---|---|---|---|---|---|
| LHet score | ||||||||
| F1 vs. F2 | 6.22 | 71.4 (41/57) | 77.8 (7/9) | 95.3 (41/43) | 30.4 (7/23) | 72.7 (48/66) | 0.845 | 0.001 |
| F2 vs. F3 | 7.02 | 53.3 (24/45) | 53.6 (30/57) | 47.1 (24/51) | 58.8 (30/51) | 52.9 (54/102) | 0.619 | 0.040 |
| F0–1 vs. F2 | 5.68 | 87.5 (50/57) | 88.0 (22/25) | 94.3 (50/53) | 75.9 (22/29) | 87.8 (72/82) | 0.944 | <0.001 |
| F0–1 vs. F3 | 6.17 | 86.7 (39/45) | 88.0 (22/25) | 92.9 (39/42) | 78.6 (22/28) | 87.1 (61/70) | 0.963 | <0.001 |
| LNod score | ||||||||
| F1 vs. F2 | 0.97 | 94.6 (54/57) | 100.0 (9/9) | 100.0 (54/54) | 75.0 (9/12) | 95.4 (63/66) | 0.958 | <0.001 |
| F2 vs. F3 | 1.12 | 64.4 (29/45) | 64.9 (37/57) | 59.2 (29/49) | 69.8 (37/53) | 64.7 (66/102) | 0.689 | 0.001 |
| F0–1 vs. F2 | 0.95 | 94.6 (54/57) | 96.0 (24/25) | 98.2 (54/55) | 88.9 (24/27) | 95.1 (78/82) | 0.969 | <0.001 |
| F0–1 vs. F3 | 0.94 | 91.1 (41/45) | 92.0 (23/25) | 95.3 (41/43) | 85.2 (23/27) | 91.4 (64/70) | 0.936 | <0.001 |
| LHet × LNod score | ||||||||
| F1 vs. F2 | 6.10 | 89.5 (51/57) | 88.9 (8/9) | 98.1 (51/52) | 57.1 (8/14) | 89.4 (59/66) | 0.954 | <0.001 |
| F2 vs. F3 | 7.85 | 66.7 (30/45) | 68.4 (39/57) | 62.5 (30/48) | 72.2 (39/54) | 67.6 (69/102) | 0.761 | <0.001 |
| F0–1 vs. F2 | 5.98 | 91.1 (52/57) | 92.0 (23/25) | 96.3 (52/54) | 82.1 (23/28) | 91.5 (75/82) | 0.984 | <0.001 |
| F0–1 vs. F3 | 6.70 | 97.8 (44/45) | 100.0 (25/25) | 100.0 (44/44) | 96.2 (25/26) | 98.6 (69/70) | 0.999 | <0.001 |
| Observer A | Observer B | p-Value * | Intrarater Reliability (ICC) † | 95% CI | p-Value † | ||
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| LHetscore | |||||||
| Overall (n = 111) | 7.06 ± 1.68 (24) | 6.94 ± 1.64 (24) | 0.140 | 0.718 | 0.589 | 0.806 | <0.001 |
| F1 (n = 9) | 5.45 ± 1.08 (20) | 5.58 ± 0.85 (15) | 0640 | 0.782 | 0.035 | 0.951 | 0.023 |
| F2 (n = 57) | 6.85 ± 1.15 (17) | 6.75 ± 1.12 (13) | 0.535 | 0.624 | 0.362 | 0.779 | <0.001 |
| F3 (n = 45) | 7.65 ± 2.05 (27) | 7.47 ± 2.08 (28) | 0.558 | 0.679 | 0.417 | 0.824 | <0.001 |
| LNodscore | |||||||
| Overall (n = 111) | 1.10 ± 0.12 (11) | 1.10 ± 0.14 (13) | 0.459 | 0.832 | 0.755 | 0.885 | <0.001 |
| F1 (n = 9) | 0.91 ± 0.04 (4) | 0.90 ± 0.06 (7) | 0.869 | 0.768 | −0.029 | 0.948 | 0.027 |
| F2 (n = 57) | 1.09 ± 0.09 (8) | 1.09 ± 0.10 (9) | 0.905 | 0.601 | 0.322 | 0.765 | <0.001 |
| F3 (n = 45) | 1.14 ± 0.13 (11) | 1.15 ± 0.16 (14) | 0.596 | 0.852 | 0.731 | 0.919 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.-H.; Jeong, C.-W.; Kim, J.E.; Kim, J.W.; Jo, H.G.; Kim, Y.R.; Lee, Y.H.; Yoon, K.-H. Assessment of Liver Fibrosis Stage Using Integrative Analysis of Hepatic Heterogeneity and Nodularity in Routine MRI with FIB-4 Index as Reference Standard. J. Clin. Med. 2021, 10, 1697. https://doi.org/10.3390/jcm10081697
Kim T-H, Jeong C-W, Kim JE, Kim JW, Jo HG, Kim YR, Lee YH, Yoon K-H. Assessment of Liver Fibrosis Stage Using Integrative Analysis of Hepatic Heterogeneity and Nodularity in Routine MRI with FIB-4 Index as Reference Standard. Journal of Clinical Medicine. 2021; 10(8):1697. https://doi.org/10.3390/jcm10081697
Chicago/Turabian StyleKim, Tae-Hoon, Chang-Won Jeong, Ji Eon Kim, Jin Woong Kim, Hoon Gil Jo, Youe Ree Kim, Young Hwan Lee, and Kwon-Ha Yoon. 2021. "Assessment of Liver Fibrosis Stage Using Integrative Analysis of Hepatic Heterogeneity and Nodularity in Routine MRI with FIB-4 Index as Reference Standard" Journal of Clinical Medicine 10, no. 8: 1697. https://doi.org/10.3390/jcm10081697






