Myeloid and T-Cell Microenvironment Immune Features Identify Two Prognostic Sub-Groups in High-Grade Gastroenteropancreatic Neuroendocrine Neoplasms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cases
2.2. Immunohistochemistry
2.3. Statistical Analysis
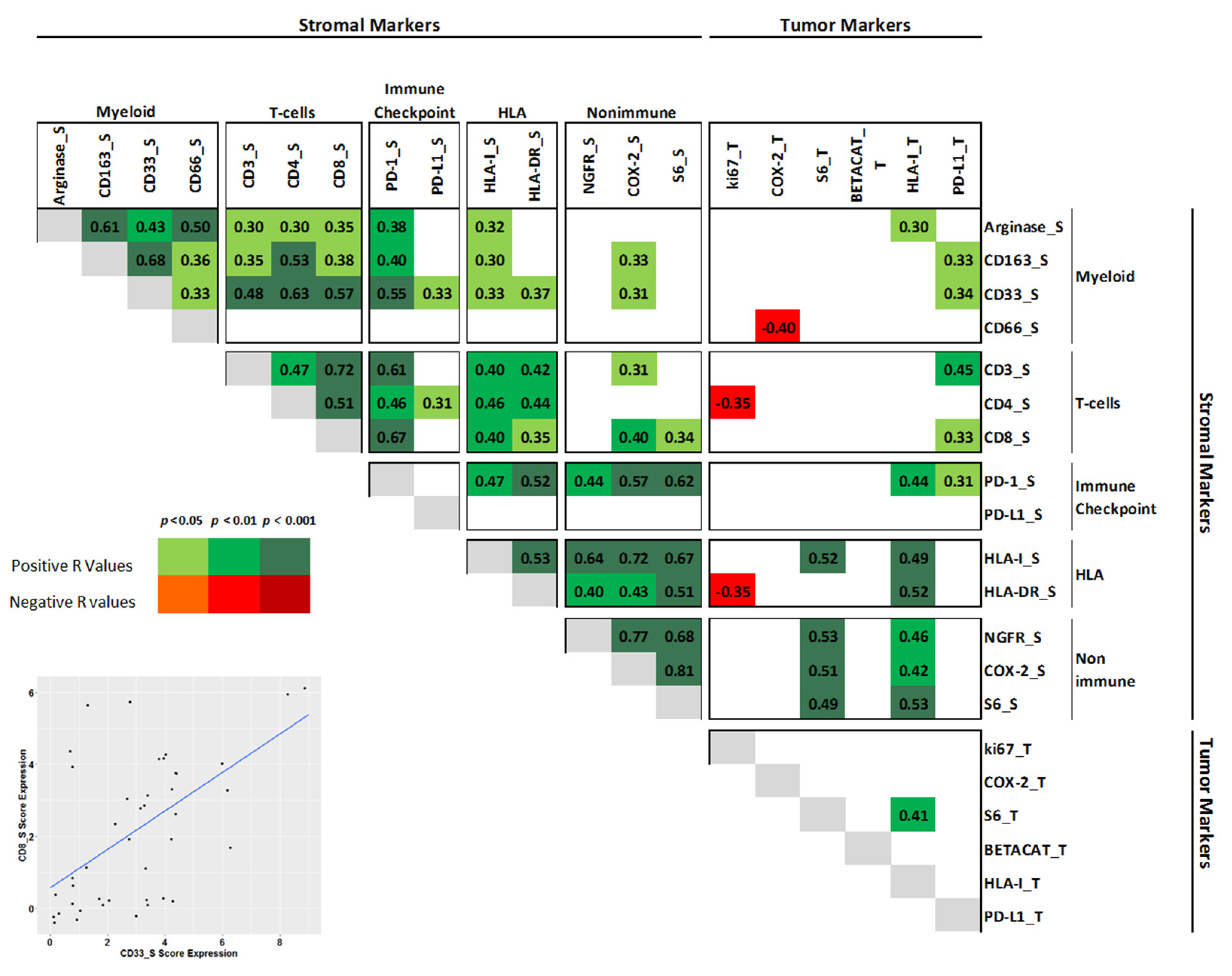
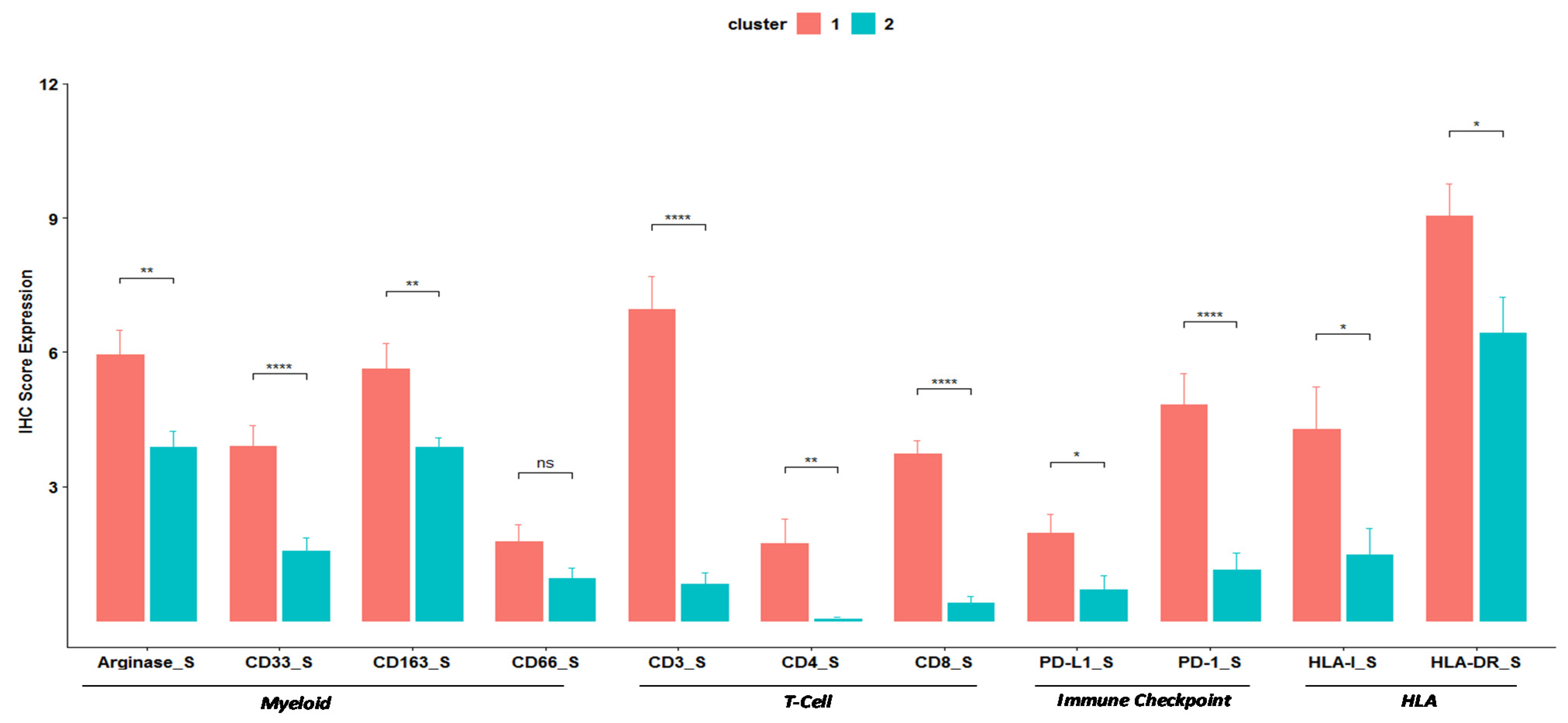
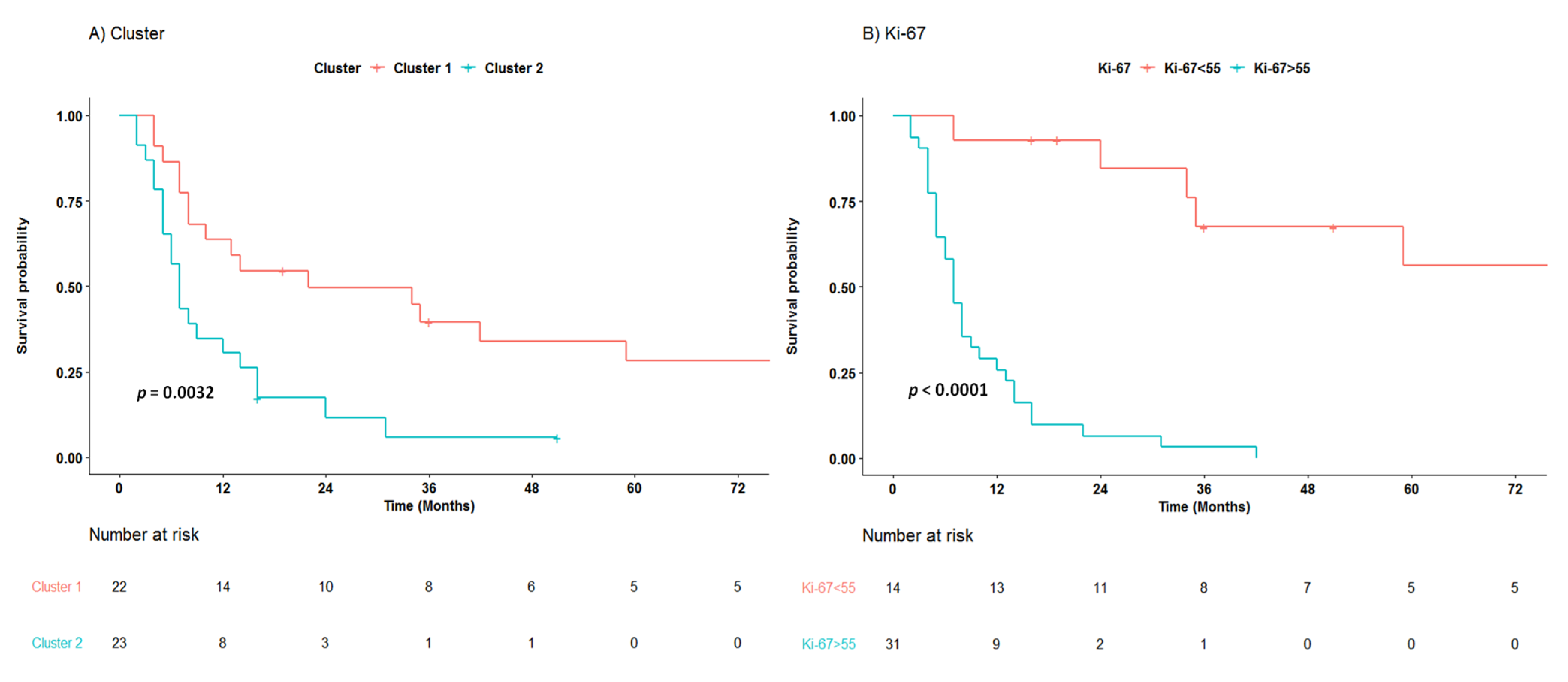
3. Results
3.1. Patient Characteristics and Myeloid Profile
3.2. Clustering
3.3. Survival Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nagtegaal, I.D.; Odze, R.D.; Klimstra, D.; Paradis, V.; Rugge, M.; Schirmacher, P.; Washington, M.K.; Carneiro, F.; Cree, I.A.; The WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020, 76, 182–188. [Google Scholar] [CrossRef] [Green Version]
- Milione, M.; Maisonneuve, P.; Spada, F.; Pellegrinelli, A.; Spaggiari, P.; Albarello, L.; Pisa, E.; Barberis, M.; Vanoli, A.; Buzzoni, R.; et al. The clinicopathologic heterogeneity of grade 3 gastroenteropancreatic neuroendocrine neoplasms: Morphological differentiation and proliferation identify different prognostic categories. Neuroendocrinology 2017, 104, 85–93. [Google Scholar] [CrossRef]
- Kiesewetter, B.; Raderer, M. How I treat neuroendocrine tumours. ESMO Open 2020, 5, e000811. [Google Scholar] [CrossRef]
- Pellat, A.; Coriat, R. Well differentiated grade 3 neuroendocrine tumors of the digestive tract: A narrative review. J. Clin. Med. 2020, 9, 1677. [Google Scholar] [CrossRef]
- Liu, A.J.; Ueberroth, B.E.; McGarrah, P.W.; Petty, S.A.B.; Kendi, A.T.; Starr, J.; Hobday, T.J.; Halfdanarson, T.R.; Sonbol, M.B. treatment outcomes of well-differentiated high-grade neuroendocrine tumors. Oncologist 2021. [Google Scholar] [CrossRef]
- Fazio, N.; Spada, F.; Giovannini, M. Chemotherapy in gastroenteropancreatic (GEP) neuroendocrine carcinomas (NEC): A critical view. Cancer Treat. Rev. 2013, 39, 270–274. [Google Scholar] [CrossRef]
- Sorbye, H.; Baudin, E.; Borbath, I.; Caplin, M.; Chen, J.; Cwikla, J.B.; Frilling, A.; Grossman, A.; Kaltsas, G.; Scarpa, A.; et al. Unmet needs in high-grade gastroenteropancreatic neuroendocrine neoplasms (WHO G3). Neuroendocrinology 2018, 108, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Garciacarbonero, R.; Sorbye, H.; Baudin, E.; Raymond, E.; Wiedenmann, B.; Niederle, B.; Sedlackova, E.; Toumpanakis, C.; Anlauf, M.; Cwikla, J.B.; et al. ENETS Consensus guidelines for high-grade gastroenteropancreatic neuroendocrine tumors and neuroendocrine carcinomas. Neuroendocrinology 2016, 103, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Klemm, F.; Joyce, J.A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015, 25, 198–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuny, T.; De Herder, W.; Barlier, A.; Hofland, L.J. Role of the tumor microenvironment in digestive neuroendocrine tumors. Endocr. Relat. Cancer 2018, 25, R519–R544. [Google Scholar] [CrossRef]
- Cai, L.; Michelakos, T.; Deshpande, V.; Arora, K.S.; Yamada, T.; Ting, D.T.; Taylor, M.S.; Castillo, C.F.-D.; Warshaw, A.L.; Lillemoe, K.D.; et al. Role of tumor associated macrophages in the clinical course of pancreatic neuroendocrine tumors (PanNETs). Clin. Cancer Res. 2019, 25, 2644–2655. [Google Scholar] [CrossRef]
- Zhang, W.-H.; Wang, W.-Q.; Gao, H.-L.; Yu, X.-J.; Liu, L. The tumor immune microenvironment in gastroenteropancreatic neuroendocrine neoplasms. Biochim. Biophys. Acta 2019, 1872, 188311. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Manuzzi, L.; Lamberti, G.; Ricci, A.D.; Tober, N.; Campana, D. Landscape and future perspectives of immunotherapy in neuroendocrine neoplasia. Cancers 2020, 12, 832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milione, M.; Miceli, R.; Barretta, F.; Pellegrinelli, A.; Spaggiari, P.; Tagliabue, G.; Centonze, G.; Paolino, C.; Mangogna, A.; Kankava, K.; et al. Microenvironment and tumor inflammatory features improve prognostic prediction in gastro-entero-pancreatic neuroendocrine neoplasms. J. Pathol. Clin. Res. 2019, 5, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milione, M.; Maisonneuve, P.; Pellegrinelli, A.; Pusceddu, S.; Centonze, G.; Dominoni, F.; Brambilla, C.; Rubino, M.; Faggiano, A.; Buzzoni, R.; et al. Loss of succinate dehydrogenase subunit B (SDHB) as a prognostic factor in advanced ileal well-differentiated neuroendocrine tumors. Endocrine 2016, 57, 512–517. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Soc. Ser. C 1979, 28, 100. [Google Scholar] [CrossRef]
- Spranger, S.; Luke, J.J.; Bao, R.; Zha, Y.; Hernandez, K.M.; Li, Y.; Gajewski, A.P.; Andrade, J.; Gajewski, T.F. Density of immunogenic antigens does not explain the presence or absence of the T-cell–inflamed tumor microenvironment in melanoma. Proc. Natl. Acad. Sci. USA 2016, 113, E7759–E7768. [Google Scholar] [CrossRef] [Green Version]
- Spranger, S.; Gajewski, T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 2018, 18, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.; Wolf, D.; Bortone, D.S.; Ouyang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The immune landscape of cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, S.C.; Donkor, C.; Glasgow, K.; Pillarisetty, V.G.; Gönen, M.; Espat, N.J.; Klimstra, D.S.; D’Angelica, M.I.; Allen, P.J.; Jarnagin, W.; et al. T cell infiltrate and outcome following resection of intermediate-grade primary neuroendocrine tumours and liver metastases. HPB 2010, 12, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, D.; Kojima, M.; Suzuki, T.; Sugimoto, M.; Kobayashi, S.; Takahashi, S.; Konishi, M.; Gotohda, N.; Ikeda, M.; Nakatsura, T.; et al. Profiling the tumour immune microenvironment in pancreatic neuroendocrine neoplasms with multispectral imaging indicates distinct subpopulation characteristics concordant with WHO 2017 Classification. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Ali, A.S.; Langer, S.W.; Federspiel, B.; Hjortland, G.O.; Grønbæk, H.; Ladekarl, M.; Welin, S.; Vestermark, L.W.; Arola, J.; Osterlund, P.; et al. PD-L1 expression in gastroenteropancreatic neuroendocrine neoplasms grade 3. PLoS ONE 2020, 15, e0243900. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Talmadge, J.E.; Gabrilovich, D.I. History of myeloid-derived suppressor cells. Nat. Rev. Cancer 2013, 13, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I. Myeloid-derived suppressor cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Zhang, Y.; Chen, L.; Lin, Y.; He, Q.; Zeng, Y.; Chen, M.; Chen, J. Myeloid-derived suppressor cells in gastroenteropancreatic neuroendocrine neoplasms. Endocrine 2021, 71, 242–252. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, X.; Zhu, C.; Liu, L.; Wang, G.; Yuan, X. the role of myeloid-derived suppressor cells in patients with solid tumors: A meta-analysis. PLoS ONE 2016, 11, e0164514. [Google Scholar] [CrossRef]
- He, T.-F.; Yost, S.E.; Frankel, P.H.; Dagis, A.; Cao, Y.; Wang, R.; Rosario, A.; Tu, T.Y.; Solomon, S.; Schmolze, D.; et al. Multi-panel immunofluorescence analysis of tumor infiltrating lymphocytes in triple negative breast cancer: Evolution of tumor immune profiles and patient prognosis. PLoS ONE 2020, 15, e0229955. [Google Scholar] [CrossRef] [Green Version]
- Kervarrec, T.; Gaboriaud, P.; Berthon, P.; Zaragoza, J.; Schrama, D.; Houben, R.; Le Corre, Y.; Hainaut-Wierzbicka, E.; Aubin, F.; Bens, G.; et al. Merkel cell carcinomas infiltrated with CD33 + myeloid cells and CD8 + T cells are associated with improved outcome. J. Am. Acad. Dermatol. 2018, 78, 973–982.e8. [Google Scholar] [CrossRef] [PubMed]
- Milione, M.; Boeri, M.; Cantarutti, A.; Centonze, G.; Busico, A.; Suatoni, P.; Garzone, G.; Cattaneo, L.; Tamborini, E.; Perrone, F.; et al. Improved prognostic prediction in never-smoker lung cancer patients by integration of a systemic inflammation marker with tumor immune contexture analysis. Cancers 2020, 12, 1828. [Google Scholar] [CrossRef] [PubMed]

| Features | All Patients |
|---|---|
| Total | 45 (100) |
| Gender | |
| M | 28 (62.2) |
| F | 17 (37.8) |
| Years | |
| Median (range) | 61 (33–78) |
| Morphology | |
| NET G3 | 6 (13.3) |
| NEC-Ki67 <55 | 8 (17.8) |
| NEC-Ki67 >55 | 31 (68.9) |
| Stage | |
| I-II | 3 (6.7) |
| III | 14 (31.1) |
| IV | 28 (62.2) |
| Site | |
| Colon | 8 (17.8) |
| Ileum | 7 (15.6) |
| Pancreas | 15 (33.3) |
| Rectum | 10 (22.2) |
| Stomach | 5 (11.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Centonze, G.; Lagano, V.; Sabella, G.; Mangogna, A.; Garzone, G.; Filugelli, M.; Belmonte, B.; Cattaneo, L.; Crisafulli, V.; Pellegrinelli, A.; et al. Myeloid and T-Cell Microenvironment Immune Features Identify Two Prognostic Sub-Groups in High-Grade Gastroenteropancreatic Neuroendocrine Neoplasms. J. Clin. Med. 2021, 10, 1741. https://doi.org/10.3390/jcm10081741
Centonze G, Lagano V, Sabella G, Mangogna A, Garzone G, Filugelli M, Belmonte B, Cattaneo L, Crisafulli V, Pellegrinelli A, et al. Myeloid and T-Cell Microenvironment Immune Features Identify Two Prognostic Sub-Groups in High-Grade Gastroenteropancreatic Neuroendocrine Neoplasms. Journal of Clinical Medicine. 2021; 10(8):1741. https://doi.org/10.3390/jcm10081741
Chicago/Turabian StyleCentonze, Giovanni, Vincenzo Lagano, Giovanna Sabella, Alessandro Mangogna, Giovanna Garzone, Martina Filugelli, Beatrice Belmonte, Laura Cattaneo, Valentina Crisafulli, Alessio Pellegrinelli, and et al. 2021. "Myeloid and T-Cell Microenvironment Immune Features Identify Two Prognostic Sub-Groups in High-Grade Gastroenteropancreatic Neuroendocrine Neoplasms" Journal of Clinical Medicine 10, no. 8: 1741. https://doi.org/10.3390/jcm10081741
APA StyleCentonze, G., Lagano, V., Sabella, G., Mangogna, A., Garzone, G., Filugelli, M., Belmonte, B., Cattaneo, L., Crisafulli, V., Pellegrinelli, A., Simbolo, M., Scarpa, A., Spaggiari, P., Brambilla, T., Pusceddu, S., Prinzi, N., Anichini, A., Tripodo, C., & Milione, M. (2021). Myeloid and T-Cell Microenvironment Immune Features Identify Two Prognostic Sub-Groups in High-Grade Gastroenteropancreatic Neuroendocrine Neoplasms. Journal of Clinical Medicine, 10(8), 1741. https://doi.org/10.3390/jcm10081741








