Cross-Sectional Exploration of Plasma Biomarkers of Alzheimer’s Disease in Down Syndrome: Early Data from the Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study
Abstract
1. Introduction
2. Experimental Section
2.1. Cognitive Assessments
2.2. Genetic Assays
Apolipoprotein E (APOE)
2.3. Biomarker Assays
2.3.1. Plasma P-tau Assays
2.3.2. NfL and 4-Plex Assays
2.4. Statistical Analyses
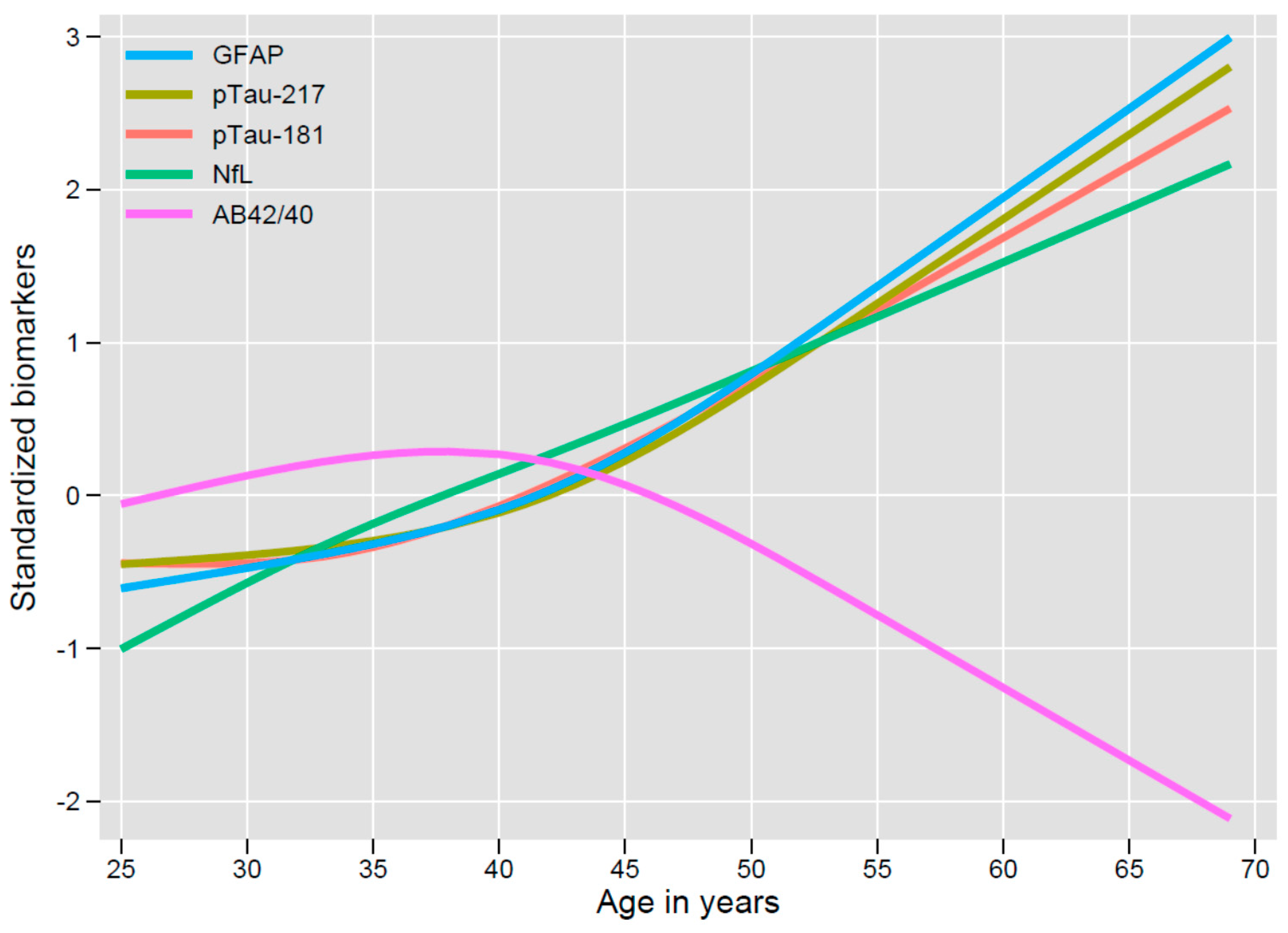
3. Results
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antonarakis, S.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, G.; Buckley, F.; Skotko, B.G. Estimation of the number of people with Down syndrome in Europe. Eur. J. Hum. Genet. 2021, 29, 402–410. [Google Scholar] [CrossRef] [PubMed]
- McCarron, M.; McCallion, P.; Reilly, E.; Dunne, P.; Carroll, R.; Mulryan, N. A prospective 20-year longitudinal follow-up of dementia in persons with Down syndrome. J. Intellect. Disabil. Res. 2017, 61, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Hithersay, R.; Startin, C.M.; Hamburg, S.; Mok, K.Y.; Hardy, J.; Fisher, E.M.C.; Tybulewicz, V.L.J.; Nizetic, D.; Strydom, A. Association of Dementia With Mortality Among Adults With Down Syndrome Older Than 35 Years. JAMA Neurol. 2019, 76, 152–160. [Google Scholar] [CrossRef]
- Lott, I.T.; Doran, E.; Nguyen, V.Q.; Tournay, A.; Movsesyan, N.; Gillen, D.L. Down syndrome and dementia: Seizures and cognitive decline. J. Alzheimers Dis. 2012, 29, 177–185. [Google Scholar] [CrossRef]
- Rissman, R.A.; Mobley, W.C. Implications for treatment: GABAA receptors in aging, Down syndrome and Alzheimer’s disease. J. Neurochem. 2011, 117, 613–622. [Google Scholar] [CrossRef]
- Alzheimer’s Association Facts and Figures. 2020. Available online: https://www.alz.org/media/Documents/alzheimers-facts-and-figures.pdf (accessed on 10 March 2020).
- Available online: https://www.alzheimers.net/resources/alzheimers-statistics/ (accessed on 8 January 2018).
- Gomez, W.; Morales, R.; Maracaja-Coutinho, V.; Parra, V.; Nassif, M. Down syndrome and Alzheimer’s disease: Common molecular traits beyond the amyloid precursor protein. Aging 2020, 12, 1011–1033. [Google Scholar] [CrossRef] [PubMed]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016, 12, 733–748. [Google Scholar] [PubMed]
- Mills, S.M.; Mallmann, J.; Santacruz, A.M.; Fuqua, A.; Carril, M.; Aisen, P.S.; Althage, M.C.; Belyew, S.; Benzinger, T.L.; Brooks, W.S.; et al. Preclinical trials in autosomal dominant AD: Implementation of the DIAN-TU trial. Rev. Neurol. 2013, 169, 737–743. [Google Scholar] [CrossRef]
- Sinai, A.; Mokrysz, C.; Bernal, J.; Bohnen, I.; Bonell, S.; Courtenay, K.; Dodd, K.; Gazizova, D.; Hassiotis, A.; Hillier, R.; et al. Predictors of Age of Diagnosis and Survival of Alzheimer’s Disease in Down Syndrome. J. Alzheimers Dis. 2018, 61, 717–728. [Google Scholar] [CrossRef]
- Ballard, C.; Mobley, W.; Hardy, J.; Williams, G.; Corbett, A. Dementia in Down’s syndrome. Lancet Neurol. 2016, 15, 622–636. [Google Scholar] [CrossRef]
- Zigman, W.B.; Devenny, D.A.; Krinsky-McHale, S.J.; Jenkins, E.C.; Urv, T.K.; Wegiel, J.; Schupf, N.; Silverman, W. Alzheimer’s Disease in Adults with Down Syndrome. Int. Rev. Res. Ment. Retard. 2008, 36, 103–145. [Google Scholar]
- Lemere, C.A.; Blusztajn, J.K.; Yamaguchi, H.; Wisniewski, T.; Saido, T.C.; Selkoe, D.J. Sequence of deposition of heterogeneous amyloid beta-peptides and APO E in Down syndrome: Implications for initial events in amyloid plaque formation. Neurobiol. Dis. 1996, 3, 16–32. [Google Scholar] [CrossRef]
- Doran, E.; Keator, D.; Head, E.; Phelan, M.J.; Kim, R.; Totoiu, M.; Barrio, J.R.; Small, G.W.; Potkin, S.G.; Lott, I.T. Down Syndrome, Partial Trisomy 21, and Absence of Alzheimer’s Disease: The Role of APP. J. Alzheimers Dis. 2017, 56, 459–470. [Google Scholar] [CrossRef]
- Liu, C.C.; Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Feldman, H.H.; Frisoni, G.B.; Hampel, H.; Jagust, W.J.; Johnson, K.A.; Knopman, D.S.; et al. An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 2016, 87, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Rafii, M.S.; Ances, B.M.; Schupf, N.; Krinsky-McHale, S.J.; Mapstone, M.; Silverman, W.; Lott, I.; Klunk, W.; Head, E.; Christian, B.; et al. The AT(N) framework for Alzheimer’s disease in adults with Down syndrome. Alzheimers Dement. 2020, 12, e12062. [Google Scholar]
- Lott, I.T.; Head, E. Dementia in Down syndrome: Unique insights for Alzheimer disease research. Nat. Rev. Neurol. 2019, 15, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Fortea, J.; Vilaplana, E.; Carmona-Iragui, M.; Benejam, B.; Videla, L.; Barroeta, I.; Fernández, S.; Altuna, M.; Pegueroles, J.; Montal, V.; et al. Clinical and biomarker changes of Alzheimer’s disease in adults with Down syndrome: A cross-sectional study. Lancet 2020, 395, 1988–1997. [Google Scholar] [CrossRef]
- Head, E.; Ances, B. Biomarkers in Down syndrome can help us understand Alzheimer’s disease. Lancet 2020, 395, 1951–1953. [Google Scholar] [CrossRef]
- Henson, R.L.; Doran, E.; Christian, B.T.; Handen, B.L.; Klunk, W.E.; Lai, F.; Lee, J.H.; Rosas, H.D.; Schupf, N.; Zaman, S.H.; et al. Cerebrospinal fluid biomarkers of Alzheimer’s disease in a cohort of adults with Down syndrome. Alzheimers Dement. 2020, 12, e12057. [Google Scholar] [CrossRef] [PubMed]
- Brickman, A.M.; Manly, J.J.; Honig, L.S.; Sanchez, D.; Reyes-Dumeyer, D.; Lantigua, R.A.; Lao, P.J.; Stern, Y.; Vonsattel, J.P.; Teich, A.F.; et al. Plasma p-tau181, p-tau217, and other blood-based Alzheimer’s disease biomarkers in a multi-ethnic, community study. Alzheimers Dement. 2021, 1–12. [Google Scholar] [CrossRef]
- Thijssen, E.H.; La Joie, R.; Wolf, A.; Strom, A.; Wang, P.; Iaccarino, L.; Bourakova, V.; Cobigo, Y.; Heuer, H.; Spina, S.; et al. Advancing Research and Treatment for Frontotemporal Lobar Degeneration (ARTFL) investigators. Diagnostic value of plasma phosphorylated tau181 in Alzheimer’s disease and frontotemporal lobar degeneration. Nat. Med. 2020, 26, 387–397. [Google Scholar] [CrossRef]
- Janelidze, S.; Mattsson, N.; Palmqvist, S.; Smith, R.; Beach, T.G.; Serrano, G.E.; Chai, X.; Proctor, N.K.; Eichenlaub, U.; Zetterberg, H.; et al. Plasma P-tau181 in Alzheimer’s disease: Relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat. Med. 2020, 26, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Fortea, J.; Carmona-Iragui, M.; Benejam, B.; Fernández, S.; Videla, L.; Barroeta, I.; Alcolea, D.; Pegueroles, J.; Muñoz, L.; Belbin, O.; et al. Plasma and CSF biomarkers for the diagnosis of Alzheimer’s disease in adults with Down syndrome: A cross-sectional study. Lancet Neurol. 2018, 17, 860–869. [Google Scholar] [CrossRef]
- Strydom, A.; Heslegrave, A.; Startin, C.M.; Mok, K.Y.; Hardy, J.; Groet, J.; Nizetic, D.; Zetterberg, H. LonDownS Consortium. Neurofilament light as a blood biomarker for neurodegeneration in Down syndrome. Alzheimers Res. Ther. 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Rabinovici, G.D.; Carrillo, M.C.; Forman, M.; DeSanti, S.; Miller, D.S.; Kozauer, N.; Petersen, R.C.; Randolph, C.; Knopman, D.S.; Smith, E.E.; et al. Multiple comorbid neuropathologies in the setting of Alzheimer’s disease neuropathology and implications for drug development. Alzheimers Dement. 2016, 3, 83–91. [Google Scholar] [CrossRef]
- Shinomoto, M.; Kasai, T.; Tatebe, H.; Kondo, M.; Ohmichi, T.; Morimoto, M.; Chiyonobu, T.; Terada, N.; Allsop, D.; Yokota, I.; et al. Plasma neurofilament light chain: A potential prognostic biomarker of dementia in adult Down syndrome patients. PLoS ONE 2019, 14, e0211575. [Google Scholar] [CrossRef]
- Verberk, I.M.W.; Thijssen, E.; Koelewijn, J.; Mauroo, K.; Vanbrabant, J.; de Wilde, A.; Zwan, M.D.; Verfaillie, S.C.J.; Ossenkoppele, R.; Barkhof, F.; et al. Combination of plasma amyloid beta(1–42/1–40) and glial fibrillary acidic protein strongly associates with cerebral amyloid pathology. Alzheimers Res. Ther. 2020, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Janelidze, S.; Quiroz, Y.T.; Zetterberg, H.; Lopera, F.; Stomrud, E.; Su, Y.; Chen, Y.; Serrano, G.E.; Leuzy, A.; et al. Discriminative Accuracy of Plasma Phospho-tau217 for Alzheimer Disease vs Other Neurodegenerative Disorders. JAMA 2020, 324, 772–781. [Google Scholar] [CrossRef] [PubMed]
- The Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study. Available online: https://clinicaltrials.gov/ct2/show/NCT04149197 (accessed on 4 November 2019).
- Fish, J. Severe Impairment Battery. In Encyclopedia of Clinical Neuropsychology; Kreutzer, J.S., DeLuca, J., Caplan, B., Eds.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Haxby, J.V. Neuropsychological evaluation of adults with Down’s syndrome: Patterns of selective impairment in non-demented old adults. J. Ment. Defic. Res. 1989, 33 Pt 3, 193–210. [Google Scholar] [CrossRef]
- Krinsky-McHale, S.J.; Zigman, W.B.; Lee, J.H.; Schupf, N.; Pang, D.; Listwan, T.; Kovacs, C.; Silverman, W. Promising outcome measures of early Alzheimer’s dementia in adults with Down syndrome. Alzheimers Dement. 2020, 12, e12044. [Google Scholar] [CrossRef] [PubMed]
- Janelidze, S.; Berron, D.; Smith, R.; Strandberg, O.; Proctor, N.K.; Dage, J.L.; Stomrud, E.; Palmqvist, S.; Mattsson-Carlgren, N.; Hansson, O. Associations of Plasma Phospho-Tau217 Levels With Tau Positron Emission Tomography in Early Alzheimer Disease. JAMA Neurol. 2021, 78, 149–156. [Google Scholar] [CrossRef]
- Janelidze, S.; Stomrud, E.; Smith, R.; Palmqvist, S.; Mattsson, N.; Airey, D.C.; Proctor, N.K.; Chai, X.; Shcherbinin, S.; Sims, J.R.; et al. Cerebrospinal fluid p-tau217 performs better than p-tau181 as a biomarker of Alzheimer’s disease. Nat. Commun. 2020, 11, 1683. [Google Scholar] [CrossRef] [PubMed]
- Mielke, M.M.; Hagen, C.E.; Xu, J.; Chai, X.; Vemuri, P.; Lowe, V.J.; Airey, D.C.; Knopman, D.S.; Roberts, R.O.; Machulda, M.M.; et al. Plasma phospho-tau181 increases with Alzheimer’s disease clinical severity and is associated with tau- and amyloid-positron emission tomography. Alzheimers Dement. 2018, 14, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Insel, P.S.; Stomrud, E.; Janelidze, S.; Zetterberg, H.; Brix, B.; Eichenlaub, U.; Dage, J.L.; Chai, X.; Blennow, K.; et al. Cerebrospinal fluid and plasma biomarker trajectories with increasing amyloid deposition in Alzheimer’s disease. EMBO Mol. Med. 2019, 11, e11170. [Google Scholar] [CrossRef]
- Hartley, S.L.; Handen, B.L.; Devenny, D.; Tudorascu, D.; Piro-Gambetti, B.; Zammit, M.D.; Laymon, C.M.; Klunk, W.E.; Zaman, S.; Cohen, A.; et al. Cognitive indicators of transition to preclinical and prodromal stages of Alzheimer’s disease in Down syndrome. Alzheimers Dement. 2020, 12, e12096. [Google Scholar] [CrossRef]
- Aschenbrenner, A.J.; Baksh, R.A.; Benejam, B.; Beresford-Webb, J.A.; Coppus, A.; Fortea, J.; Handen, B.L.; Hartley, S.; Head, E.; Jaeger, J.; et al. Markers of early changes in cognition across cohorts of adults with Down syndrome at risk of Alzheimer’s disease. Alzheimers Dement. 2021, in press. [Google Scholar]
| Characteristic | Mean (SD) | Median (Q1, Q3) | Min, Max | N (%) |
|---|---|---|---|---|
| p-tau 217 pg/mL | 0.26 (0.38) | 0.14 (0.08, 0.25) | 0.01, 2.72 | 90 (100.00%) |
| p-tau 181 pg/mL | 1.22 (1.10) | 0.81 (0.59, 1.45) | 0.37, 7.40 | 90 (100.00%) |
| NfL pg/mL | 13.84 (7.19) | 11.90 (9.28, 18.06) | 5.13, 46.87 | 90 (100.00%) |
| GFAP pg/mL | 100.37 (57.02) | 84.74 (63.44, 123.65) | 36.19, 445.03 | 90 (100.00%) |
| Aβ1-40 pg/mL | 210.03 (52.36) | 215.82 (188.07, 234.87) | 66.32, 417.74 | 90 (100.00%) |
| Aβ1-42 pg/mL | 8.81 (2.42) | 8.90 (7.36, 10.24) | 2.22, 13.82 | 90 (100.00%) |
| Aβ42/Aβ40 ratio | 0.04 (0.01) | 0.04 (0.04, 0.05) | 0.01, 0.06 | 90 (100.00%) |
| DS-MSE Total Score 2 | 64.23 (11.25) | 65.00 (59.00, 72.00) | 30.00, 82.00 | 79 (87.78%) |
| SIB Total Score | 87.92 (14.04) | 93.00 (86.00, 96.50) | 14.00, 100.00 | 84 (93.33%) |
| Age (years) | 38.31 (9.47) | 37.00 (30.00, 45.00) | 25.00, 69.00 | 86 (95.56%) |
| Female | Male | |||
| Gender | 40 (46.51%) | 46 (53.49%) | ||
| Non-Carrier | Carrier | |||
| APOE ε4 | 67 (74.44%) | 23 (25.56%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hendrix, J.A.; Airey, D.C.; Britton, A.; Burke, A.D.; Capone, G.T.; Chavez, R.; Chen, J.; Chicoine, B.; Costa, A.C.S.; Dage, J.L.; et al. Cross-Sectional Exploration of Plasma Biomarkers of Alzheimer’s Disease in Down Syndrome: Early Data from the Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study. J. Clin. Med. 2021, 10, 1907. https://doi.org/10.3390/jcm10091907
Hendrix JA, Airey DC, Britton A, Burke AD, Capone GT, Chavez R, Chen J, Chicoine B, Costa ACS, Dage JL, et al. Cross-Sectional Exploration of Plasma Biomarkers of Alzheimer’s Disease in Down Syndrome: Early Data from the Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study. Journal of Clinical Medicine. 2021; 10(9):1907. https://doi.org/10.3390/jcm10091907
Chicago/Turabian StyleHendrix, James A., David C. Airey, Angela Britton, Anna D. Burke, George T. Capone, Ronelyn Chavez, Jacqueline Chen, Brian Chicoine, Alberto C. S. Costa, Jeffrey L. Dage, and et al. 2021. "Cross-Sectional Exploration of Plasma Biomarkers of Alzheimer’s Disease in Down Syndrome: Early Data from the Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study" Journal of Clinical Medicine 10, no. 9: 1907. https://doi.org/10.3390/jcm10091907
APA StyleHendrix, J. A., Airey, D. C., Britton, A., Burke, A. D., Capone, G. T., Chavez, R., Chen, J., Chicoine, B., Costa, A. C. S., Dage, J. L., Doran, E., Esbensen, A., Evans, C. L., Faber, K. M., Foroud, T. M., Hart, S., Haugen, K., Head, E., Hendrix, S., ... Mobley, W. (2021). Cross-Sectional Exploration of Plasma Biomarkers of Alzheimer’s Disease in Down Syndrome: Early Data from the Longitudinal Investigation for Enhancing Down Syndrome Research (LIFE-DSR) Study. Journal of Clinical Medicine, 10(9), 1907. https://doi.org/10.3390/jcm10091907







