Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms
Abstract
1. Introduction
2. Methods
2.1. Study Subjects
2.2. Target SNP and Genotyping
2.3. Data Analysis
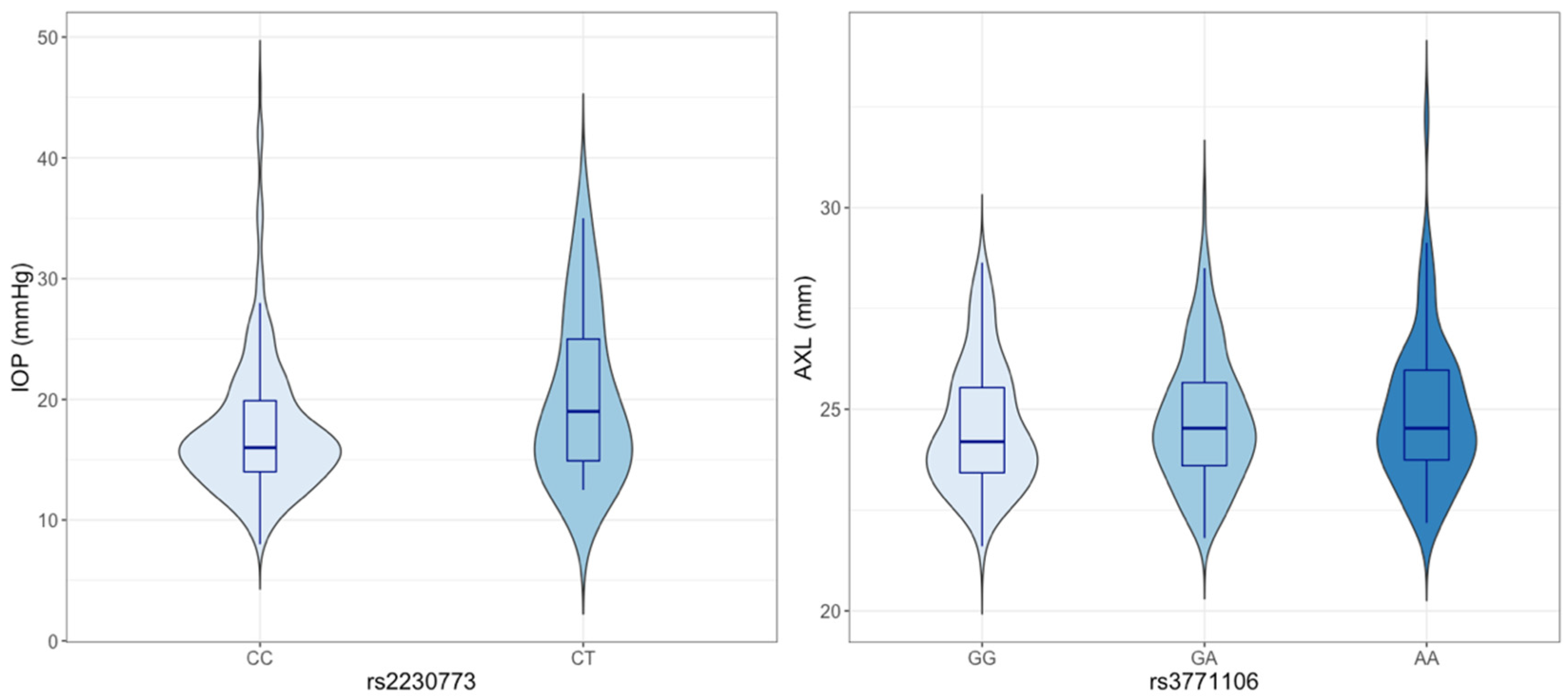
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARHGEF12 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AXL | axial length |
| CCT | central corneal thickness |
| C/D | cup-to-disc ratio |
| GCIPL | ganglion cell-inner plexiform layer |
| GLAU-GENDISK | GLAUcoma GENe DIscovery Study in Korea |
| HTG | high-tension glaucoma |
| IOP | intraocular pressure |
| LIM | Lin-11/Isl-1/Mec-3 |
| MD | mean deviation |
| MLC | myosin light chain |
| NTG | normal-tension glaucoma |
| OCT | optical coherence tomography |
| OR | odds ratio |
| POAG | primary open-angle glaucoma |
| RGC | retinal ganglion cells |
| RNFL | retinal nerve fiber layer |
| ROCK | Rho-associated coiled-coil kinase |
| SNP | single-nucleotide polymorphism |
| TM | trabecular meshwork |
| VF | visual field |
References
- Leung, T.; Manser, E.; Tan, L.; Lim, L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem. 1995, 270, 29051–29054. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, O.; Fujisawa, K.; Ishizaki, T.; Saito, Y.; Nakao, K.; Narumiya, S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996, 392, 189–193. [Google Scholar] [CrossRef]
- Inoue, T.; Tanihara, H. Rho-associated kinase inhibitors: A novel glaucoma therapy. Prog. Retin. Eye Res. 2013, 37, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Fukata, Y.; Kaibuchi, K. Regulation and functions of Rho-associated kinase. Exp. Cell Res. 2000, 261, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Julian, L.; Olson, M.F. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small Gtpases 2014, 5, e29846. [Google Scholar] [CrossRef] [PubMed]
- Moura-Coelho, N.; Tavares Ferreira, J.; Bruxelas, C.P.; Dutra-Medeiros, M.; Cunha, J.P.; Pinto Proenca, R. Rho kinase inhibitors-a review on the physiology and clinical use in Ophthalmology. Graefe’s Arch. Clin. Exp. Ophthalmol. Albrecht Von Graefes Arch. Fur Klin. Und Exp. Ophthalmol. 2019, 257, 1101–1117. [Google Scholar] [CrossRef] [PubMed]
- Tanna, A.P.; Johnson, M. Rho Kinase Inhibitors as a Novel Treatment for Glaucoma and Ocular Hypertension. Ophthalmology 2018, 125, 1741–1756. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.K.; Mack, H.; Teusch, N. Rho kinase, a promising drug target for neurological disorders. Nat. Rev. Drug Discov. 2005, 4, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Tokushige, H.; Waki, M.; Takayama, Y.; Tanihara, H. Effects of Y-39983, a selective Rho-associated protein kinase inhibitor, on blood flow in optic nerve head in rabbits and axonal regeneration of retinal ganglion cells in rats. Curr. Eye Res. 2011, 36, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Chihara, E.; Dimitrova, G.; Chihara, T. Increase in the OCT angiographic peripapillary vessel density by ROCK inhibitor ripasudil instillation: A comparison with brimonidine. Graefe’s Arch. Clin. Exp. Ophthalmol. Albrecht Von Graefes Arch. Fur Klin. Und Exp. Ophthalmol. 2018, 256, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Kim, Y.J.; Cheong, H.S.; Shiga, Y.; Hashimoto, K.; Song, Y.J.; Kim, S.H.; Choi, H.J.; Nishiguchi, K.M.; Kawai, Y.; et al. Exploring the Novel Susceptibility Gene Variants for Primary Open-Angle Glaucoma in East Asian Cohorts: The GLAU-GENDISK Study. Sci. Rep. 2020, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Loirand, G. Rho Kinases in Health and Disease: From Basic Science to Translational Research. Pharm. Rev. 2015, 67, 1074–1095. [Google Scholar] [CrossRef] [PubMed]
- Demiryurek, S.; Okumus, S.; Bozgeyik, I.; Oztuzcu, S.; Coskun, E.; Mat, E.; Durucu, E.; Tatar, M.G.; Erbagci, I.; Gurler, B.; et al. Investigation of the Rho-kinase Gene Polymorphism in Primary Open-angle Glaucoma. Ophthalmic Genet. 2016, 37, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Palomino Doza, J.; Topf, A.; Bentham, J.; Bhattacharya, S.; Cosgrove, C.; Brook, J.D.; Granados-Riveron, J.; Bu’Lock, F.A.; O’Sullivan, J.; Stuart, A.G.; et al. Low-frequency intermediate penetrance variants in the ROCK1 gene predispose to Tetralogy of Fallot. BMC Genet. 2013, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Zee, R.Y.L.; Wang, Q.M.; Chasman, D.I.; Ridker, P.M.; Liao, J.K. Gene variations of ROCKs and risk of ischaemic stroke: The Women’s Genome Health Study. Clin. Sci. 2014, 126, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Tabur, S.; Oztuzcu, S.; Oguz, E.; Korkmaz, H.; Eroglu, S.; Ozkaya, M.; Demiryurek, A.T. Association of Rho/Rho-kinase gene polymorphisms and expressions with obesity-related metabolic syndrome. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1680–1688. [Google Scholar] [PubMed]
- Zhao, R.; Liu, K.; Huang, Z.; Wang, J.; Pan, Y.; Huang, Y.; Deng, X.; Liu, J.; Qin, C.; Cheng, G.; et al. Genetic Variants in Caveolin-1 and RhoA/ROCK1 Are Associated with Clear Cell Renal Cell Carcinoma Risk in a Chinese Population. PLoS ONE 2015, 10, e0128771. [Google Scholar] [CrossRef] [PubMed]
- Pehlivan, Y.; Yolbas, S.; Cetin, G.Y.; Alibaz-Oner, F.; Cagatay, Y.; Yilmaz, N.; Oztuzcu, S.; Donmez, S.; Ozgen, M.; Koca, S.S.; et al. Investigation of the association between Rho/Rho-kinase gene polymorphisms and systemic sclerosis. Rheumatol. Int. 2016, 36, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Springelkamp, H.; Iglesias, A.I.; Cuellar-Partida, G.; Amin, N.; Burdon, K.P.; van Leeuwen, E.M.; Gharahkhani, P.; Mishra, A.; van der Lee, S.J.; Hewitt, A.W.; et al. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum. Mol. Genet. 2015, 24, 2689–2699. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.K.; Kee, C. Population-based glaucoma prevalence studies in Asians. Surv. Ophthalmol. 2014, 59, 434–447. [Google Scholar] [CrossRef] [PubMed]
| N | ID | Tagging SNP (r2 = 1) | Gene | Chr | Position | Location | AA Change |
|---|---|---|---|---|---|---|---|
| 1 | rs2271621 | rs2290156 | ROCK2 | 2 | 11193868 | intron | |
| 2 | rs34945852 | ROCK2 | 2 | 11197557 | missense | K997M | |
| 3 | rs202027620 | ROCK2 | 2 | 11200967 | missense | T881I | |
| 4 | rs35768389 | ROCK2 | 2 | 11214974 | missense | D515V | |
| 5 | rs190769228 | ROCK2 | 2 | 11215516 | missense | N445H | |
| 6 | rs2230773 | ROCK2 | 2 | 11249688 | synonymous | ||
| 7 | rs3771106 | rs2230774, rs1515219, rs726843, rs978906 | ROCK2 | 2 | 11249986 | intron | |
| 8 | rs965665 | ROCK2 | 2 | 11258749 | intron | ||
| 9 | rs10178332 | ROCK2 | 2 | 11268891 | intron | ||
| 10 | rs1130157 | ROCK2 | 2 | 11286615 | 5’UTR | ||
| 11 | rs6755196 | ROCK2 | 2 | 11320199 | intron | ||
| 12 | rs10929732 | ROCK2 | 2 | 11343366 | intron | ||
| 13 | rs75122528 | ROCK1 | 18 | 20947821 | 3’UTR | ||
| 14 | rs2847081 | ROCK1 | 18 | 20948500 | 3’UTR | ||
| 15 | rs8089184 | ROCK1 | 18 | 20970650 | intron | ||
| 16 | rs288980 | ROCK1 | 18 | 21029619 | intron | ||
| 17 | rs288979 | rs7239317 | ROCK1 | 18 | 21031282 | intron | |
| 18 | rs2127958 | ROCK1 | 18 | 21073649 | intron | ||
| 19 | rs1481280 | ROCK1 | 18 | 21075490 | intron | ||
| 20 | rs1006881 | rs11874761 | ROCK1 | 18 | 21101332 | intron | |
| 21 | rs35996865 | ROCK1 | 18 | 21112383 | 5’near | ||
| 22 | rs10083915 | ROCK1 | 18 | 21120994 | 5’near | ||
| 23 | rs11873284 | ROCK1 | 18 | 21135030 | 5’near | ||
| 24 | rs1515210 | ROCK1 | 18 | 29283684 | 5’ |
| Variable | POAG (n = 363) | Healthy (n = 213) | p-Value | |
| Age, year | 54.0 ± 13.7 | 54.6 ± 9.7 | 0.55 * | |
| NTG | 54.3 ± 13.3 | 54.6 ± 9.7 | 0.76 * | |
| HTG | 53.1 ± 15.3 | 54.6 ± 9.7 | 0.40 * | |
| Sex (Female), n (%) | 180 (49.6%) | 93 (43.7%) | 0.17 † | |
| NTG | 155 (55.0%) | 93 (43.7%) | 0.016 † | |
| HTG | 25 (30.9%) | 93 (43.7%) | 0.06 † | |
| NTG (n = 282) | HTG (n = 81) | |||
| Baseline IOP, mm Hg | 15.3 ± 3.0 | 26.0 ± 6.5 | <0.001 * | |
| CCT, µm | 532.9 ± 35.9 | 546.9 ± 30.2 | 0.001 * | |
| AXL, mm | 24.7 ± 1.6 | 24.7 ± 1.8 | 0.96 * | |
| DH history | 36 (12.8%) | 3 (3.7%) | 0.034† | |
| Rim area, mm2 | 0.77 ± 0.20 | 0.67 ± 0.29 | 0.013 * | |
| Disc area, mm2 | 1.95 ± 0.50 | 1.93 ± 0.40 | 0.80 * | |
| Vertical C/D | 0.77 ± 0.11 | 0.81 ± 0.11 | 0.024 * | |
| Average RNFL thickness, μM | 70.9 ± 11.6 | 64.3 ± 14.3 | 0.001 * | |
| Average GCIPL thickness, μM | 67.9 ± 8.8 | 63.5 ± 11.0 | 0.004 * | |
| MD of VF, dB | −8.4 ± 6.4 | −14.5 ± 10.0 | <0.001 * | |
| Gene | ID | Alleles | POAG | |||
|---|---|---|---|---|---|---|
| Case MAF | Control MAF | OR (95%CI) | p-Value | |||
| ROCK2 | rs2271621 | G > T | 0.486 | 0.479 | 1.03 (0.81, 1.32) | 0.80 |
| ROCK2 | rs202027620 | G > A | 0.000 | 0.005 | ||
| ROCK2 | rs2230773 | C > T | 0.024 | 0.024 | 1.04 (0.47, 2.33) | 0.92 |
| ROCK2 | rs3771106 | G > A | 0.448 | 0.433 | 1.06 (0.83, 1.36) | 0.63 |
| ROCK2 | rs965665 | G > C | 0.008 | 0.002 | 3.69 (0.44, 31.01) | 0.17 |
| ROCK2 | rs10178332 | A > C | 0.008 | 0.002 | 3.69 (0.44, 31.01) | 0.17 |
| ROCK2 | rs6755196 | G > A | 0.008 | 0.002 | 3.68 (0.44, 30.94) | 0.17 |
| ROCK2 | rs10929732 | G > A | 0.036 | 0.045 | 0.80 (0.44, 1.47) | 0.48 |
| ROCK1 | rs75122528 | A > T | 0.300 | 0.319 | 0.92 (0.71, 1.19) | 0.54 |
| ROCK1 | rs2847081 | T > C | 0.138 | 0.127 | 1.09 (0.76, 1.56) | 0.65 |
| ROCK1 | rs8089184 | T > C | 0.145 | 0.131 | 1.11 (0.78, 1.58) | 0.56 |
| ROCK1 | rs288980 | T > C | 0.475 | 0.474 | 1.00 (0.79, 1.27) | 0.99 |
| ROCK1 | rs288979 | A > G | 0.138 | 0.129 | 1.08 (0.76, 1.54) | 0.67 |
| ROCK1 | rs2127958 | T > C | 0.441 | 0.445 | 0.99 (0.78, 1.25) | 0.90 |
| ROCK1 | rs1481280 | C > A | 0.300 | 0.312 | 0.95 (0.74, 1.23) | 0.72 |
| ROCK1 | rs1006881 | G > A | 0.140 | 0.129 | 1.10 (0.77, 1.56) | 0.61 |
| ROCK1 | rs35996865 | T > G | 0.138 | 0.129 | 1.08 (0.76, 1.54) | 0.68 |
| ROCK1 | rs10083915 | A > G | 0.141 | 0.129 | 1.10 (0.77, 1.56) | 0.60 |
| ROCK1 | rs11873284 | A > G | 0.141 | 0.129 | 1.10 (0.77, 1.57) | 0.60 |
| ROCK1 | rs1515210 | C > G | 0.183 | 0.181 | 1.03 (0.75, 1.41) | 0.86 |
| Gene | ID | Alleles | Control MAF | NTG | HTG | ||||
|---|---|---|---|---|---|---|---|---|---|
| Case MAF | OR (95%CI) | p-Value | Case MAF | OR (95%CI) | p-Value | ||||
| ROCK2 | rs2271621 | G > T | 0.479 | 0.491 | 1.05 (0.81, 1.36) | 0.71 | 0.468 | 0.96 (0.66, 1.42) | 0.85 |
| ROCK2 | rs202027620 | G > A | 0.005 | 0.000 | 0.000 | ||||
| ROCK2 | rs2230773 | C > T | 0.024 | 0.020 | 0.83 (0.34, 2.00) | 0.67 | 0.039 | 1.59 (0.55, 4.56) | 0.40 |
| ROCK2 | rs3771106 | G > A | 0.433 | 0.453 | 1.09 (0.84, 1.41) | 0.54 | 0.429 | 1.00 (0.67, 1.47) | 0.99 |
| ROCK2 | rs965665 | G > C | 0.002 | 0.011 | 4.94 (0.58, 41.83) | 0.09 | 0.000 | ||
| ROCK2 | rs10178332 | A > C | 0.002 | 0.011 | 4.94 (0.58, 41.83) | 0.09 | 0.000 | ||
| ROCK2 | rs6755196 | G > A | 0.002 | 0.011 | 4.92 (0.58, 41.72) | 0.09 | 0.000 | ||
| ROCK2 | rs10929732 | G > A | 0.045 | 0.036 | 0.78 (0.41, 1.49) | 0.45 | 0.039 | 0.83 (0.33, 2.09) | 0.69 |
| ROCK1 | rs75122528 | A > T | 0.319 | 0.282 | 0.86 (0.65, 1.13) | 0.27 | 0.364 | 1.19 (0.82, 1.74) | 0.36 |
| ROCK1 | rs2847081 | T > C | 0.127 | 0.153 | 1.22 (0.84, 1.77) | 0.28 | 0.076 | 0.56 (0.28, 1.14) | 0.09 |
| ROCK1 | rs8089184 | T > C | 0.131 | 0.164 | 1.28 (0.89, 1.84) | 0.18 | 0.078 | 0.56 (0.29, 1.08) | 0.07 |
| ROCK1 | rs288980 | T > C | 0.474 | 0.475 | 1.00 (0.78, 1.29) | 1.00 | 0.474 | 0.99 (0.69, 1.42) | 0.97 |
| ROCK1 | rs288979 | A > G | 0.129 | 0.157 | 1.26 (0.87, 1.82) | 0.22 | 0.071 | 0.52 (0.26, 1.03) | 0.045 |
| ROCK1 | rs2127958 | T > C | 0.445 | 0.441 | 0.99 (0.77, 1.28) | 0.95 | 0.442 | 0.98 (0.68, 1.40) | 0.90 |
| ROCK1 | rs1481280 | C > A | 0.312 | 0.280 | 0.88 (0.67, 1.15) | 0.34 | 0.370 | 1.25 (0.87, 1.82) | 0.23 |
| ROCK1 | rs1006881 | G > A | 0.129 | 0.159 | 1.28 (0.89, 1.84) | 0.19 | 0.071 | 0.52 (0.26, 1.03) | 0.045 |
| ROCK1 | rs35996865 | T > G | 0.129 | 0.156 | 1.26 (0.87, 1.82) | 0.22 | 0.071 | 0.52 (0.26, 1.03) | 0.045 |
| ROCK1 | rs10083915 | A > G | 0.129 | 0.161 | 1.28 (0.89, 1.83) | 0.19 | 0.071 | 0.52 (0.26, 1.03) | 0.045 |
| ROCK1 | rs11873284 | A > G | 0.129 | 0.160 | 1.28 (0.89, 1.85) | 0.18 | 0.071 | 0.52 (0.26, 1.03) | 0.045 |
| ROCK1 | rs1515210 | C > G | 0.181 | 0.185 | 1.05 (0.75, 1.46) | 0.78 | 0.175 | 0.94 (0.58, 1.52) | 0.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-W.; Bak, E.; Wy, S.; Lee, S.-C.; Kim, Y.-J.; Kim, Y.-K.; Park, K.-H.; Jeoung, J.-W. Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms. J. Clin. Med. 2021, 10, 1953. https://doi.org/10.3390/jcm10091953
Kim Y-W, Bak E, Wy S, Lee S-C, Kim Y-J, Kim Y-K, Park K-H, Jeoung J-W. Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms. Journal of Clinical Medicine. 2021; 10(9):1953. https://doi.org/10.3390/jcm10091953
Chicago/Turabian StyleKim, Yong-Woo, Eunoo Bak, Seoyoung Wy, Seung-Chan Lee, Yu-Jeong Kim, Young-Kook Kim, Ki-Ho Park, and Jin-Wook Jeoung. 2021. "Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms" Journal of Clinical Medicine 10, no. 9: 1953. https://doi.org/10.3390/jcm10091953
APA StyleKim, Y.-W., Bak, E., Wy, S., Lee, S.-C., Kim, Y.-J., Kim, Y.-K., Park, K.-H., & Jeoung, J.-W. (2021). Genetic Risk and Phenotype Correlation of Primary Open-Angle Glaucoma Based on Rho-Kinase Gene Polymorphisms. Journal of Clinical Medicine, 10(9), 1953. https://doi.org/10.3390/jcm10091953







