A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19
Abstract
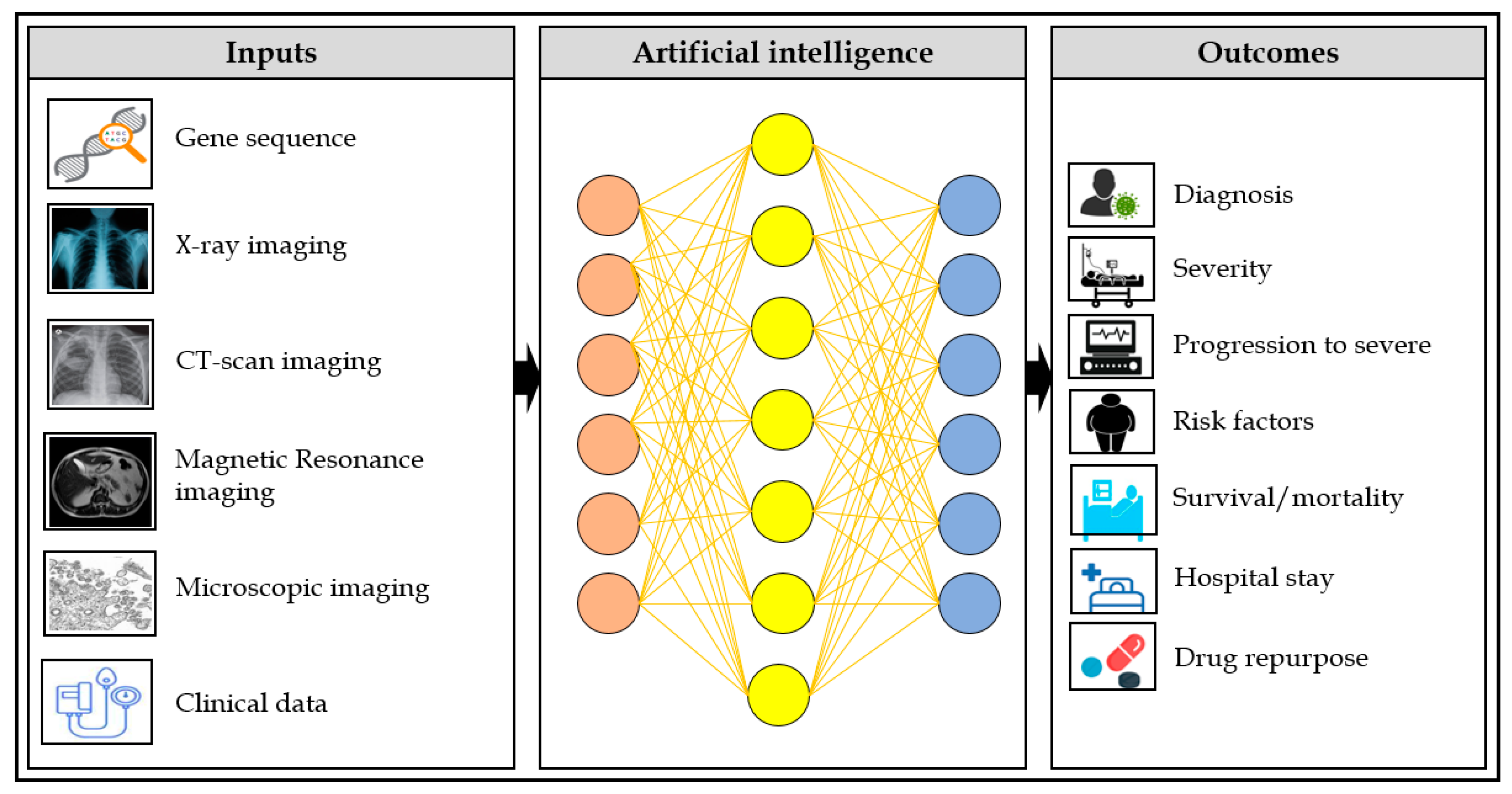
1. Introduction
2. Overview of Artificial Intelligence
2.1. Machine Learning
2.1.1. Random Forest (RF)
2.1.2. Support Vector Machine (SVM)
2.1.3. Logistic Regression (LR)
2.1.4. XGBoost
2.2. Deep Learning
2.2.1. Artificial Neural Networks (ANNs)
2.2.2. Convolutional Neural Network (CNN)
2.2.3. Neural Recurrent Network (RNN)
2.2.4. Long Short Term Memory (LSTM)
3. Review of State-of-the-Art
3.1. COVID-19 Screening Using Digital Images
3.1.1. Potentiality
3.1.2. Limitations
3.2. Artificial Intelligence for COVID-19 Severity
3.2.1. Potentiality
3.2.2. Limitations
3.3. Artificial Intelligence for COVID-19 Mortality
3.3.1. Potentiality
3.3.2. Limitations
3.4. Artificial Intelligence for COVID-19 Drug Repurposing
3.4.1. Potentiality
3.4.2. Limitation
3.5. Artificial Intelligence for Epidemic Trends
4. Overall Challenges to Deploy AI Model in the Clinical Settings
- The number of participants used to train the AI models to predict disease progression, mortality risk was not sufficient to deploy in real-world clinical settings. It is a great challenge to train the model using a large number of patients from multiple sites or countries and make the AI model more generable and trustworthy;
- As all of the studies used different types (laboratory, symptoms, biochemical, CT/X-ray) and a various number of variables to predict the risk of severity and mortality; therefore, it is challenging to establish what kinds of variables should be used, and what the optimal number to be utilized is while admitting COVID-19 patient to the hospital. The traditional scoring systems for stratifying patients have a fixed number of variables, but deciding the fixed number of variables from those studies may be difficult;
- Making strong evidence and the simplicity of prediction models is also challenging to fight against COVID-19. All of the included studies used different data sets, and the ethnicity was also different. Moreover, they reported a different time frame while predicting disease progression and mortality risk. All of the studies should provide a standard time frame, such as 24 h, 3 days, and 7 days to predict patient’s situation;
- Generalizability is another potential challenge to deploy the AI model in the real-world clinical setting to tackle COVID-19. The findings of one study might be different while testing it using other datasets from different countries;
- Reducing bias, such as patient selection, reference standard, and methodology, would be challenging. However, all of the upcoming studies should follow standard guidelines (e.g., TRIPOD) while reporting their findings;
- Resolving the “black-box” issue would be more challenging; however, all of the studies should provide a clear explanation of predictors and how these predictors influence the performance. They should report univariate and multivariate analysis while showing the performance metrics. Moreover, they should categorize the variables (e.g., symptoms, laboratory, and radiology) and present the model performance for each category;
- Others (Recommendation from organizations, establishing trust among healthcare providers, decreasing false positive and negative results, and ethical issues).
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, F.; Deng, Y.; Li, W. Coronavirus disease 2019: What we know? J. Med. Virol. 2020, 92, 719–725. [Google Scholar] [CrossRef]
- Woo, P.C.; Huang, Y.; Lau, S.K.; Yuen, K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef]
- Zu, Z.Y.; Jiang, M.D.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus disease 2019 (COVID-19): A perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Norrie, J.D. Remdesivir for COVID-19: Challenges of underpowered studies. Lancet 2020, 395, 1525–1527. [Google Scholar] [CrossRef]
- Mitjà, O.; Clotet, B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob. Health 2020, 8, e639–e640. [Google Scholar] [CrossRef]
- Tufan, A.; Güler, A.A.; Matucci-Cerinic, M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk. J. Med. Sci. 2020, 50, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Kai, H.; Kai, M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020, 43, 648–654. [Google Scholar] [CrossRef]
- Willner, P.; Rose, J.; Stenfert Kroese, B.; Murphy, G.H.; Langdon, P.E.; Clifford, C.; Hutchings, H.; Watkins, A.; Hiles, S.; Cooper, V. Effect of the COVID-19 pandemic on the mental health of carers of people with intellectual disabilities. J. Appl. Res. Intellect. Disabil. 2020, 33, 1523–1533. [Google Scholar] [CrossRef]
- Walsh, E.E.; Falsey, A.R.; Swinburne, I.A.; Formica, M.A. Reverse transcription polymerase chain reaction (RT-PCR) for diagnosis of respiratory syncytial virus infection in adults: Use of a single-tube “hanging droplet” nested PCR. J. Med. Virol. 2001, 63, 259–263. [Google Scholar] [CrossRef]
- Kim, H.; Hong, H.; Yoon, S.H. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: A meta-analysis. Radiology 2020, 296, E145–E155. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.K. Bootstrapping text recognition from stop words. In Proceedings of the Fourteenth International Conference on Pattern Recognition (Cat No 98EX170), Brisbane, QLD, Australia, 20 August 1998; pp. 605–609. [Google Scholar]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Oshiro, T.M.; Perez, P.S.; Baranauskas, J.A. How many trees in a random forest? In Proceedings of the International Workshop on Machine Learning and Data Mining in Pattern Recognition, Berlin, Germany, 13–20 July 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 154–168. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning; Springer Series in Statistics: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Suthaharan, S. Machine learning models and algorithms for big data classification. Integr. Ser. Inf. Syst. 2016, 36, 1–12. [Google Scholar]
- Kumar, R.; Arora, R.; Bansal, V.; Sahayasheela, V.J.; Buckchash, H.; Imran, J.; Narayanan, N.; Pandian, G.N.; Raman, B. Accurate prediction of COVID-19 using chest X-ray images through deep feature learning model with smote and machine learning classifiers. MedRxiv 2020. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.; Döhmen, T.; Rebholz-Schuhmann, D.; Decker, S.; Cochez, M.; Beyan, O. Deepcovidexplainer: Explainable covid-19 predictions based on chest X-ray images. arXiv 2020, arXiv:200404582. [Google Scholar]
- Hemdan, E.E.-D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv 2020, arXiv:200311055. [Google Scholar]
- Civit-Masot, J.; Luna-Perejón, F.; Domínguez Morales, M.; Civit, A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl. Sci. 2020, 10, 4640. [Google Scholar] [CrossRef]
- Elaziz, M.A.; Hosny, K.M.; Salah, A.; Darwish, M.M.; Lu, S.; Sahlol, A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE 2020, 15, e0235187. [Google Scholar] [CrossRef]
- Wang, D.; Mo, J.; Zhou, G.; Xu, L.; Liu, Y. An efficient mixture of deep and machine learning models for COVID-19 diagnosis in chest X-ray images. PLoS ONE 2020, 15, e0242535. [Google Scholar] [CrossRef]
- Das, A.K.; Kalam, S.; Kumar, C.; Sinha, D. TLCoV-An automated Covid-19 screening model using Transfer Learning from chest X-ray images. Chaos Solitons Fractals 2021, 144, 110713. [Google Scholar] [CrossRef] [PubMed]
- Kassani, S.H.; Kassasni, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Automatic detection of coronavirus disease (covid-19) in X-ray and ct images: A machine learning-based approach. arXiv 2020, arXiv:200410641. [Google Scholar]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Jain, G.; Mittal, D.; Thakur, D.; Mittal, M.K. A deep learning approach to detect Covid-19 coronavirus with X-ray images. Biocybern. Biomed. Eng. 2020, 40, 1391–1405. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Kumar, V.; Kaur, M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Panigrahi, B.K.; Dey, N.; Rajinikanth, V.; Gandhi, T.K. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl. Intell. 2021, 51, 571–585. [Google Scholar] [CrossRef]
- Docherty, A.B.; Harrison, E.M.; Green, C.A.; Hardwick, H.E.; Pius, R.; Norman, L.; Holden, K.A.; Read, J.M.; Dondelinger, F.; Carson, G. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Prospective observational cohort study. BMJ 2020, 369. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; Zhang, Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef]
- Cai, W.; Liu, T.; Xue, X.; Luo, G.; Wang, X.; Shen, Y.; Fang, Q.; Sheng, J.; Chen, F.; Liang, T. CT Quantification and Machine-learning Models for Assessment of Disease Severity and Prognosis of COVID-19 Patients. Acad. Radiol. 2020, 27, 1665–1678. [Google Scholar] [CrossRef]
- Lassau, N.; Ammari, S.; Chouzenoux, E.; Gortais, H.; Herent, P.; Devilder, M.; Soliman, S.; Meyrignac, O.; Talabard, M.-P.; Lamarque, J.-P. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Yip, S.S.; Klanecek, Z.; Naganawa, S.; Kim, J.; Studen, A.; Rivetti, L.; Jeraj, R. Performance and Robustness of Machine Learning-based Radiomic COVID-19 Severity Prediction. medRxiv 2020. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Zha, Y. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, J.C.; Feng, Y.-Z.; Cheng, Z.-Y.; Rezazadegan, D.; Chen, P.-K.; Lin, Q.-T.; Qian, L.; Liu, X.-F.; Berkovsky, S.; Coiera, E. Development and validation of a machine learning approach for automated severity assessment of COVID-19 based on clinical and imaging data: Retrospective study. JMIR Med. Inform. 2021, 9, e24572. [Google Scholar] [CrossRef] [PubMed]
- Aktar, S.; Ahamad, M.; Rashed-Al-Mahfuz, M.; Azad, A.; Uddin, S.; Kamal, A.; Alyami, S.A.; Lin, P.-I.; Islam, S.M.S.; Quinn, J.M. Predicting Patient COVID-19 Disease Severity by means of Statistical and Machine Learning Analysis of Blood Cell Transcriptome Data. arXiv 2020, arXiv:201110657. [Google Scholar]
- Feng, Y.; Liu, S.; Cheng, Z.; Quiroz, J.; Chen, P.; Lin, Q.; Qian, L.; Liu, X.; Berkovsky, S.; Coiera, E. Severity Assessment and Progression Prediction of COVID-19 Patients based on the LesionEncoder Framework and Chest CT. medRxiv 2020. [Google Scholar] [CrossRef]
- Xiao, L.-S.; Li, P.; Sun, F.; Zhang, Y.; Xu, C.; Zhu, H.; Cai, F.-Q.; He, Y.-L.; Zhang, W.-F.; Ma, S.-C. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of Coronavirus disease 2019. Front. Bioeng. Biotechnol. 2020, 8, 898. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yang, P.; Xie, Y.; Woodruff, H.C.; Rao, X.; Guiot, J.; Frix, A.-N.; Louis, R.; Moutschen, M.; Li, J. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: An international multicentre study. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Q.; Tan, Y.; Feng, X.; Yue, Y.; Bai, Y.; Li, J.; Li, J.; Xu, Y.; Chen, S. Prediction of COVID-19 Severity Using Chest Computed Tomography and Laboratory Measurements: Evaluation Using a Machine Learning Approach. JMIR Med. Inform. 2020, 8, e21604. [Google Scholar] [CrossRef]
- Kang, J.; Chen, T.; Luo, H.; Luo, Y.; Du, G.; Jiming-Yang, M. Machine learning predictive model for severe COVID-19. Infect. Genet. Evol. 2021, 90, 104737. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.T.; Park, J.; Kim, T.; Park, B.; Lee, J.; Kim, J.Y.; Kim, K.B.; Choi, S.; Kim, Y.H.; Lim, J.-K. Deep Learning Models for Predicting Severe Progression in COVID-19-Infected Patients: Retrospective Study. JMIR Med. Inform. 2021, 9, e24973. [Google Scholar] [CrossRef] [PubMed]
- Pourhomayoun, M.; Shakibi, M. Predicting mortality risk in patients with COVID-19 using artificial intelligence to help medical decision-making. medRxiv 2020. [Google Scholar] [CrossRef]
- Bertsimas, D.; Lukin, G.; Mingardi, L.; Nohadani, O.; Orfanoudaki, A.; Stellato, B.; Wiberg, H.; Gonzalez-Garcia, S.; Parra-Calderon, C.L.; Robinson, K. COVID-19 mortality risk assessment: An international multi-center study. PLoS ONE 2020, 15, e0243262. [Google Scholar] [CrossRef]
- Malki, Z.; Atlam, E.-S.; Hassanien, A.E.; Dagnew, G.; Elhosseini, M.A.; Gad, I. Association between weather data and COVID-19 pandemic predicting mortality rate: Machine learning approaches. Chaos Solitons Fractals 2020, 138, 110137. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.L.; Abels, E.; McCaffrey, P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021, 34, 522–531. [Google Scholar] [CrossRef]
- Zhou, B.; She, J.; Wang, Y.; Ma, X. Utility of ferritin, procalcitonin, and C-reactive protein in severe patients with 2019 novel coronavirus disease. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Poly, T.N.; Islam, M.M.; Yang, H.C.; Lin, M.C.; Jian, W.-S.; Hsu, M.-H.; Li, Y.-C.J. Obesity and Mortality Among Patients Diagnosed With COVID-19: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8. [Google Scholar] [CrossRef]
- Mehra, M.R.; Desai, S.S.; Kuy, S.; Henry, T.D.; Patel, A.N. Cardiovascular disease, drug therapy, and mortality in Covid-19. N. Engl. J. Med. 2020, 382, e102. [Google Scholar] [CrossRef] [PubMed]
- Abdulaal, A.; Patel, A.; Charani, E.; Denny, S.; Mughal, N.; Moore, L. Prognostic modeling of COVID-19 using artificial intelligence in the United Kingdom: Model development and validation. J. Med. Internet Res. 2020, 22, e20259. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Lim, H.; Kim, D.-W.; Chang, J.H.; Choi, Y.J.; Kim, S.W. Machine learning prediction for mortality of patients diagnosed with COVID-19: A nationwide Korean cohort study. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, G.-Y.; Fang, W.; Li, H.-Y.; Wang, S.-Y.; Chen, L.; Yu, Y.; Liu, D.; Xu, S.; Cui, P.-F. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Liu, Z.; Jiang, Y.; Shi, O.; Zhang, X.; Xu, K.; Suo, C.; Wang, Q.; Song, Y.; Yu, K. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int. J. Epidemiol. 2020, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ge, P.; Zhu, J.; Li, H.; Graham, J.; Singer, A.; Richman, P.S.; Duong, T.Q. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ 2020, 8, e10337. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, H.-T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Rechtman, E.; Curtin, P.; Navarro, E.; Nirenberg, S.; Horton, M.K. Vital signs assessed in initial clinical encounters predict COVID-19 mortality in an NYC hospital system. Sci. Rep. 2020, 10, 1–6. [Google Scholar] [CrossRef]
- Ryan, L.; Lam, C.; Mataraso, S.; Allen, A.; Green-Saxena, A.; Pellegrini, E.; Hoffman, J.; Barton, C.; McCoy, A.; Das, R. Mortality prediction model for the triage of COVID-19, pneumonia, and mechanically ventilated ICU patients: A retrospective study. Ann. Med. Surg. 2020, 59, 207–216. [Google Scholar] [CrossRef]
- Vaid, A.; Somani, S.; Russak, A.J.; De Freitas, J.K.; Chaudhry, F.F.; Paranjpe, I.; Johnson, K.W.; Lee, S.J.; Miotto, R.; Richter, F. Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation. J. Med. Internet Res. 2020, 22, e24018. [Google Scholar] [CrossRef]
- Yadaw, A.S.; Li, Y.-C.; Bose, S.; Iyengar, R.; Bunyavanich, S.; Pandey, G. Clinical features of COVID-19 mortality: Development and validation of a clinical prediction model. Lancet Digit. Health 2020, 2, e516–e525. [Google Scholar] [CrossRef]
- Adams, C.P.; Brantner, V.V. Estimating the cost of new drug development: Is it really $802 million? Health Aff. 2006, 25, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Persidis, A. The benefits of drug repositioning. Drug Discov. World 2011, 12, 9–12. [Google Scholar]
- Cantürk, S.; Singh, A.; St-Amant, P.; Behrmann, J. Machine-learning driven drug repurposing for covid-19. arXiv 2020, arXiv:200614707. [Google Scholar]
- Choi, Y.; Shin, B.; Kang, K.; Park, S.; Beck, B.R. Target-Centered Drug Repurposing Predictions of Human Angiotensin-Converting Enzyme 2 (ACE2) and Transmembrane Protease Serine Subtype 2 (TMPRSS2) Interacting Approved Drugs for Coronavirus Disease 2019 (COVID-19) Treatment through a Drug-Target Interaction Deep Learning Model. Viruses 2020, 12, 1325. [Google Scholar]
- Esmail, S.; Danter, W. Viral pandemic preparedness: A pluripotent stem cell-based machine-learning platform for simulating SARS-CoV-2 infection to enable drug discovery and repurposing. Stem Cells Transl. Med. 2021, 10, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, S.; Stoyanov, S.R. COSMO-RS-Based Descriptors for the Machine Learning-Enabled Screening of Nucleotide Analogue Drugs against SARS-CoV-2. J. Phys. Chem. Lett. 2020, 11, 9408–9414. [Google Scholar] [CrossRef] [PubMed]
- Hooshmand, S.A.; Ghobadi, M.Z.; Hooshmand, S.E.; Jamalkandi, S.A.; Alavi, S.M.; Masoudi-Nejad, A. A multimodal deep learning-based drug repurposing approach for treatment of COVID-19. Mol. Divers. 2020, 1–14. [Google Scholar] [CrossRef]
- Ioannidis, V.N.; Zheng, D.; Karypis, G. Few-shot link prediction via graph neural networks for covid-19 drug-repurposing. arXiv 2020, arXiv:200710261. [Google Scholar]
- Ke, Y.-Y.; Peng, T.-T.; Yeh, T.-K.; Huang, W.-Z.; Chang, S.-E.; Wu, S.-H.; Hung, H.-C.; Hsu, T.-A.; Lee, S.-J.; Song, J.-S. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020, 43, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kowalewski, J.; Ray, A. Predicting novel drugs for SARS-CoV-2 using machine learning from a >10 million chemical space. Heliyon 2020, 6, e04639. [Google Scholar] [CrossRef] [PubMed]
- Loucera, C.; Esteban-Medina, M.; Rian, K.; Falco, M.M.; Dopazo, J.; Peña-Chilet, M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal. Transduct. Target. Ther. 2020, 5, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Nath, P.; Chatterjee, M.; Das, N.; Kalita, D.; Roy, P.; Satapathi, S. Repurposing therapeutics for COVID-19: Rapid prediction of commercially available drugs through machine learning and docking. PLoS ONE 2020, 15, e0241543. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.-H.; Qiu, Y.; Zeng, J.; Xie, L.; Zhang, P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat. Mach. Intell. 2021, 3, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Aggarwal, R. Repurposing potential of FDA approved and investigational drugs for COVID-19 targeting SARS-CoV-2 spike and main protease and validation by machine learning algorithm. Chem. Biol. Drug Des. 2020, 97. [Google Scholar] [CrossRef]
- Zeng, X.; Song, X.; Ma, T.; Pan, X.; Zhou, Y.; Hou, Y.; Zhang, Z.; Li, K.; Karypis, G.; Cheng, F. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 2020, 19, 4624–4636. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zheng, X.; Ai, G.; Liu, D.; Zhu, B. Time series prediction for the epidemic trends of COVID-19 using the improved LSTM deep learning method: Case studies in Russia, Peru and Iran. Chaos Solitons Fractals 2020, 140, 110214. [Google Scholar] [CrossRef]






| Author | Model | Algorithms | Applications | Modality | F-1 Score | AUROC/Accuracy |
|---|---|---|---|---|---|---|
| Hemdan [20] | CNN | DenseNet | Classification of COVID-19 and normal | X-ray | 0.91 | - |
| Civit-Masot [21] | CNN | VGG16 | Classification of COVID-19, Pneumonia, and healthy | X-ray | 0.91 | >90 |
| Elaziz [22] | CNN | FrMEMs | Classification of COVID-19 and healthy | X-ray | - | --/96 and 98 |
| Wang [23] | CNN | Xception + SVM | Classification of COVID-19 and normal | X-ray | - | 99.33/99.32 |
| Das [24] | CNN | VGG-16 | Classification of COVID-19, Pneumonia and normal | X-ray | 0.96 | --/97.67 |
| Kassani [25] | CNN | DesnseNet121+Bagging | Classification of COVID-19 and normal | X-ray and CT scan | 0.96 | --/99 |
| Ardakani [26] | CNN | ResNet-101 | Classification of COVID-19 and normal | CT scan | 1.0 | 99.4/99.5 |
| Jain [27] | CNN | ResNet101 | Classification of COVID-19 and viral pneumonia | X-ray | 0.98 | --/98.15 |
| Singh [28] | CNN | MODE-based CNN | Classification of COVID-19 and normal | CT scan | -- | --/93.3 |
| Ahuja [29] | CNN | ResNet 18 | Classification of COVID-19 and normal | CT scan | 0.99 | 99.65/99.4 |
| Author | Methods | Application | Variable Types | Precision/Recall | AUROC/Accuracy |
|---|---|---|---|---|---|
| Akbar [37] | GBM | Severity of COVID-19 | Blood | 0.91/0.88 | 89/89 |
| Feng [38] | RNN | Severity | CT scan | --/0.81 | 90/94 |
| Xiao [39] | CNN | Severity | CT scan | --/-- | 89/81.9 |
| Wu [40] | LR | Severity | CT and laboratory | 0.66~0.95/ 0.75~0.96 | 84~93/ 74.4~87.5 |
| Li [41] | CNN | Severity | CT and laboratory | 0.82/0.79 | 93/88 |
| Kang [42] | ANN | Severity | CT, clinical and laboratory | --/-- | 95/-- |
| Ho [43] | CNN | Severity | CT | 0.78/0.80 | 91/93 |
| Author | Methods | Application | Variable | Sensitivity/Specificity | AUROC/Accuracy |
|---|---|---|---|---|---|
| Abdulaal [52] | ANN | Mortality risk | Demographic, comorbidities, smoking history, and symptom | 0.87/0.85 | -/86.25 |
| An [53] | SVM | Mortality risk | Demographics, symptom, comorbidities, and medications | 0.92/0.91 | 96.3/- |
| Gao [54] | Ensemble model | Mortality risk | Demographics, comorbidity and vital sign | 0.32~0.45/ 0.97~0.99 | 92~97/ 93.0~95.6 |
| Hu [55] | LR | Mortality risk | Demographic and laboratory | 0.83/0.79 | 88/- |
| Li [56] | ANN | Mortality risk | Demographics, symptoms and laboratory | 0.75/0.87 | 84/85 |
| Yan [57] | XGBoost | Mortality risk | Demographic, symptom, and laboratory | 1/- | 92.2~95.05/ |
| Rechtman [58] | XGBoost | Mortality risk | Demographics, symptoms, comorbidities | - | 86/- |
| Ryan [59] | XGBoost | Mortality risk | Demographic, comorbidity, vital sign, and laboratory | 0.82/0.84 | 91.0/80 |
| Vaid [60] | XGBoost | Mortality risk | Demographic, comorbidity, vital sign, and laboratory | - | 68~98/ |
| Yadaw [61] | XGBoost | Mortality risk | Demographics, comorbidity, smoking | - | -/91 |
| Author | Application | Model | Data | Results |
|---|---|---|---|---|
| Beck-2020 [4] | Identifying available drugs that could act on viral proteins of SARS-CoV-2 using Molecule Transformer-Drug Target Interaction (MT-DTI) | Transfer learning and molecular docking | Drug Target Common (DTC) database and BindingDB | antiviral drugs such as lopinavir/ritonavir had been identified by the MT-DTI model should be considered |
| Choi-2020 [65] | Finding approved drugs that can inhibit COVID-19 by using g a deep learning-based drug-target interaction model called Molecule Transformer-Drug Target Interaction (MT-DTI) | Transfer learning and molecular docking | DrugBank and ZINC | Identified 30 drugs that have strong inhibitory potencies to the angiotensin converting Enzyme 2 (ACE2) receptor and the transmembrane protease serine 2 (TMPRSS2). |
| Esmail-2020 [66] | Identifying antiviral therapeutic targets for drug repurposing by using the DeepNEU stem cell-based platform and validated computer simulations of artificial lung cells. | Hybrid deep-machine learning system with elements of fully connected RNNs, CMs, and evolutionary systems (GA) | DeepNEU database plus important information upgrades in the form of a new gene, protein, and phenotypic relationship data. | To improve preparedness for and response to future viral outbreaks. |
| Gusarov-2020 [67] | Identifying potential drugs for SARS-CoV-2 using machine learning algorithms | Machine learning algorithms | N/A | Short for conductor-like screening model for real solvents might assist to accelerate drug discovery for the treatment of COVID-19 |
| Hooshmand-2020 [68] | Finding potential drugs that can inhibit COVID-19 using the Multimodal Restricted Boltzmann Machine approach (MM-RBM) | Multimodal Restricted Boltzmann Machine approach (MM-RBM) | Harmonizome and Literacy Information and Communication System (LINCS) | MM-RBM has immense potential to identify the highly promising medications for COVID-19 with minimum side effects. |
| N. Ioannidis-2020 [69] | Identifying COVID-19 drugs for repurposing using deep graph learning | RGCN and state-of-the-art KGE | IMDB, DBLP and drug-repurposing knowledge-graph (DRKG) | Their model showed promise to identify possible drug candidates. |
| Ke-2020 [70] | Identifying the marketed drugs with potential for treating COVID-19 using artificial intelligence method | Deep Neural Network (DNN) | DrugBank, | Identified 80 potential drugs that have the ability to fight coronavirus. |
| Kowalewski-2020 [71] | Searching several drug candidates for COVID-19 using machine learning algorithms. | Support vector machine | ZINC, ChEMBL 25, DrugBank, EPI Suite, Therapeutic targets databases, Hazardous substances data Bank | Suggested several drugs for repurposed that suited for short-term approval, and long-term approval need follow-up |
| Loucera-2020 [72] | Aimed at using machine learning models to identify appropriate drugs fight against SARS-CoV-2 infection | Machine learning | DrugBank | It shows promising results and found several drugs that can be considered only a subset of the potential drug candidates for repurposing. |
| Mohapatra-2020 [73] | Developed a machine-learning model to find drugs already available in the market; can be used for inhibiting SARS-CoV-2 infection. | Classification models such as Naïve Bayes, molecular docking | PubChem Bioassay, DrugBank | The findings suggested that machine-learning algorithms can be identified and tested the therapeutic agents for COVID-19 treatment. |
| Pham-2020 [74] | Identifying strong associations among biological features, and outputs to predict gene expression profiles given a new chemical compound. | DeepCE based on linear models, vanilla neural network, k-nearest neighbor, and tensor-train weight optimization models. | L1000 gene expression gene, STRING, DrugBank, Gene Expression Omnibus | DeepCE helps to accelerate compound screening against a single target. |
| Verma-2020 [75] | To evaluate potential response of existing antiviral drug candidates against SARS-CoV-2 target proteins that help viral entry and replication into the host body. | Bayesian machine learning | PubChem, ZINC, DrugBank, | Their model identified 45 drugs that can inhibit SARS-CoV-2. Those drugs work on the major target proteins such as spike protein (S) and main proteases. |
| Zeng-2020 [76] | To develop a network-based deep-learning method of identifying drugs to work as repurpose drugs for COVID-19 | DGL-KE developed by AWS AI | PubMed, DrugBank | Their model identified 41 repurpose drugs that may accelerate therapeutic response against COVID-19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.M.; Poly, T.N.; Alsinglawi, B.; Lin, M.C.; Hsu, M.-H.; Li, Y.-C. A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19. J. Clin. Med. 2021, 10, 1961. https://doi.org/10.3390/jcm10091961
Islam MM, Poly TN, Alsinglawi B, Lin MC, Hsu M-H, Li Y-C. A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19. Journal of Clinical Medicine. 2021; 10(9):1961. https://doi.org/10.3390/jcm10091961
Chicago/Turabian StyleIslam, Md. Mohaimenul, Tahmina Nasrin Poly, Belal Alsinglawi, Ming Chin Lin, Min-Huei Hsu, and Yu-Chuan (Jack) Li. 2021. "A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19" Journal of Clinical Medicine 10, no. 9: 1961. https://doi.org/10.3390/jcm10091961
APA StyleIslam, M. M., Poly, T. N., Alsinglawi, B., Lin, M. C., Hsu, M.-H., & Li, Y.-C. (2021). A State-of-the-Art Survey on Artificial Intelligence to Fight COVID-19. Journal of Clinical Medicine, 10(9), 1961. https://doi.org/10.3390/jcm10091961








