Potential Contribution of IL-27 and IL-23 Gene Polymorphisms to Multiple Sclerosis Susceptibility: An Association Analysis at Genotype and Haplotype Level
Abstract
:1. Introduction
2. Materials and Methods
2.1. Methods
2.1.1. DNA Extraction
2.1.2. Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP Analysis)
2.2. Statistical Analysis
3. Results
3.1. Study Subjects
3.2. Association Analysis between the Studied SNPs and Odds of MS
3.3. SNP-SNP Interactions between IL-27-T4730, IL-27-A964G and IL-23-R381 and MS Risk
3.4. Analysis of Haplotypes and Linkage Disequilibrium (LD) between IL-27-T4730, IL-27-A964G SNPs
3.5. Association Analysis of MS Characteristics with SNPs of IL-27-T4730, IL-27-A964G and IL-23-R381Q
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wildner, P.; Stasiołek, M.; Matysiak, M. Differential diagnosis of multiple sclerosis and other inflammatory CNS diseases. Mult. Scler. Relat. Disord. 2020, 37, 101452. [Google Scholar] [CrossRef] [PubMed]
- Number of people with MS|Atlas of MS. Available online: https://www.atlasofms.org/map/global/epidemiology/number-of-people-with-ms (accessed on 14 October 2020).
- Zeydan, B.; Kantarci, O.H. Progressive Forms of Multiple Sclerosis: Distinct Entity or Age-Dependent Phenomena. Neurol. Clin. 2018, 36, 163–171. [Google Scholar] [CrossRef]
- Thompson, A.J.; Baranzini, S.E.; Geurts, J.; Hemmer, B.; Ciccarelli, O. Multiple sclerosis. Lancet 2018, 391, 1622–1636. [Google Scholar] [CrossRef]
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple Sclerosis: Pathogenesis, Symptoms, Diagnoses and Cell-Based Therapy. Cell J. 2017, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Trojano, M.; Avolio, C. Environmental Factors and Their Regulation of Immunity in Multiple Sclerosis. Transl. Neuroimmunol. Mult. Scler. Dis. Mech. Clin. Appl. 2016, 324, 100–111. [Google Scholar] [CrossRef]
- Iluţ, S.; Nistor, C.; Nemes, B.; Dudea, M.S. Therapeutic difficulties in vegetative epileptic seizures as a sign of acute viral encephalitis: Case presentation. Balneo Res. J. 2020, 29–34. [Google Scholar] [CrossRef]
- Silva, B.A.; Leal, M.C.; Farías, M.I.; Avalos, J.C.; Besada, C.H.; Pitossi, F.J.; Ferrari, C.C. A new focal model resembling features of cortical pathology of the progressive forms of multiple sclerosis: Influence of innate immunity. Brain. Behav. Immun. 2018, 69, 515–531. [Google Scholar] [CrossRef]
- Correale, J.; Gaitán, M.I.; Ysrraelit, M.C.; Fiol, M.P. Progressive multiple sclerosis: From pathogenic mechanisms to treatment. Brain 2017, 140, 527–546. [Google Scholar] [CrossRef] [Green Version]
- Oksenberg, J.R.; McCauley, J.L. Genetics of Multiple Sclerosis. In Translational Neuroimmunology in Multiple Sclerosis; Elsevier: Amsterdam, The Netherlands, 2016; pp. 45–54. ISBN 9780128020074. [Google Scholar]
- Hemmer, B.; Kerschensteiner, M.; Korn, T. Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 2015, 14, 406–419. [Google Scholar] [CrossRef]
- Hossain, M.J.; Tanasescu, R.; Gran, B. Innate immune regulation of autoimmunity in multiple sclerosis: Focus on the role of Toll-like receptor 2. J. Neuroimmunol. 2017, 304, 11–20. [Google Scholar] [CrossRef] [PubMed]
- De Jager, P.L.; Chibnik, L.B.; Cui, J.; Lehr, S.; Simon, K.C. Multiple Sclerosis Susceptibility. Integrating 2011, 8, 1111–1119. [Google Scholar] [CrossRef] [Green Version]
- Gourraud, P.-A.; McElroy, J.P.; Caillier, S.J.; Johnson, B.A.; Santaniello, A.; Hauser, S.L.; Oksenberg, J.R. Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann. Neurol. 2011, 69, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, J.; Kim, D.; Kim, S.; Valentin-Torres, A.; Dvorina, N.; Jang, E.; Nagarajavel, V.; Desilva, T.M.; Li, X.; Ting, A.H.; et al. Treg-specific IL-27Rα deletion uncovers a key role for IL-27 in Treg function to control autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, 10190–10195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behzadi, P.; Behzadi, E.; Ranjbar, R. Il-12 family cytokines: General characteristics, pathogenic microorganisms, receptors, and signalling pathways. Acta Microbiol. Immunol. Hung. 2016, 63, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.B.S. Clinical Immunology, 5th ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 1, ISBN 9780702068966. [Google Scholar]
- Sivanesan, D.; Beauchamp, C.; Quinou, C.; Lee, J.; Lesage, S.; Chemtob, S.; Rioux, J.D.; Michnick, S.W. IL23R (Interleukin 23 Receptor) variants protective against inflammatory bowel diseases (IBD) display loss of function due to impaired protein stability and intracellular trafficking. J. Biol. Chem. 2016, 291, 8673–8685. [Google Scholar] [CrossRef] [Green Version]
- Langrish, C.L.; Chen, Y.; Blumenschein, W.M.; Mattson, J.; Basham, B.; Sedgwick, J.D.; McClanahan, T.; Kastelein, R.A.; Cua, D.J. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005, 201, 233–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reboldi, A.; Coisne, C.; Baumjohann, D.; Benvenuto, F.; Bottinelli, D.; Lira, S.; Uccelli, A.; Lanzavecchia, A.; Engelhardt, B.; Sallusto, F. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat. Immunol. 2009, 10, 514–523. [Google Scholar] [CrossRef]
- Yu, R.Y.; Brazaitis, J.; Gallagher, G. The Human IL-23 Receptor rs11209026 A Allele Promotes the Expression of a Soluble IL-23R–Encoding mRNA Species. J. Immunol. 2015, 194, 1062–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, J.; Rueda, B.; López-Nevot, M.A.; Gómez-García, M.; Martín, J. Replication of an Association Between IL23R Gene Polymorphism With Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. 2007, 5, 977–981.e2. [Google Scholar] [CrossRef]
- Sáfrány, E.; Pazár, B.; Csöngei, V.; Járomi, L.; Polgár, N.; Sipeky, C.; Horváth, I.F.; Zeher, M.; Poór, G.; Melegh, B. Variants of the IL23R gene are associated with ankylosing spondylitis but not with sjögren syndrome in hungarian population samples. Scand. J. Immunol. 2009, 70, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Pidasheva, S.; Trifari, S.; Phillips, A.; Hackney, J.A.; Ma, Y.; Smith, A.; Sohn, S.J.; Spits, H.; Little, R.D.; Behrens, T.W.; et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS ONE 2011, 6, e25038. [Google Scholar] [CrossRef] [Green Version]
- Gyülvészi, G.; Haak, S.; Becher, B. IL-23-driven encephalo-tropism and Th17 polarization during CNS-inflammation in vivo. Eur. J. Immunol. 2009, 39, 1864–1869. [Google Scholar] [CrossRef]
- Brucklacher-Waldert, V.; Stuerner, K.; Kolster, M.; Wolthausen, J.; Tolosa, E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain 2009, 132, 3329–3341. [Google Scholar] [CrossRef] [PubMed]
- Hasheminia, S.J.; Tolouei, S.; Zarkesh-Esfahani, S.H.; Shaygannejad, V.; Shirzad, H.A.; Torabi, R.; Hashem Zadeh Chaloshtory, M. Cytokines gene expression in newly diagnosed multiple sclerosis patients. Iran. J. Allergy. Asthma. Immunol. 2015, 14, 208–216. [Google Scholar] [PubMed]
- Lu, D.; Lu, J.; Ji, X.; Ji, Y.; Zhang, Z.; Peng, H.; Sun, F.; Zhang, C. IL-27 suppresses airway inflammation, hyperresponsiveness and remodeling via the STAT1 and STAT3 pathways in mice with allergic asthma. Int. J. Mol. Med. 2020, 46, 641–652. [Google Scholar] [CrossRef]
- Kalliolias, G.D.; Ivashkiv, L.B. IL-27 Activates Human Monocytes via STAT1 and Suppresses IL-10 Production but the Inflammatory Functions of IL-27 Are Abrogated by TLRs and p38. J. Immunol. 2008, 180, 6325–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.X.; Yu, C.R.; Mahdi, R.M.; Egwuagu, C.E. Novel IL27p28/IL12p40 cytokine suppressed experimental autoimmune uveitis by inhibiting autoreactive Th1/Th17 cells and promoting expansion of regulatory T cells. J. Biol. Chem. 2012, 287, 36012–36021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, E.S. Transcriptional regulation of T helper 17 cell differentiation. Yonsei Med. J. 2010, 51, 484–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawanokuchi, J.; Takeuchi, H.; Sonobe, Y.; Mizuno, T.; Suzumura, A. Interleukin-27 promotes inflammatory and neuroprotective responses in microglia. Clin. Exp. Neuroimmunol. 2013, 4, 36–45. [Google Scholar] [CrossRef]
- Tüzün, E. Immunopathological Factors Associated with Disability in Multiple Sclerosis. Noro Psikiyatr. Ars. 2018, 55, S26–S30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhang, P.; Tang, T.; Liao, H.; Zhang, K.; Pu, Y.; Chen, P.; Song, Y.; Zhang, L. Polymorphisms and plasma levels of IL-27: Impact on genetic susceptibility and clinical outcome of bladder cancer. BMC Cancer 2015, 15, 433. [Google Scholar] [CrossRef] [Green Version]
- Jahantigh, D.; Zidanloo, S.G.; Forghani, F.; Doroudian, M. IL-27 variants might be genetic risk factors for preeclampsia: Based on genetic polymorphisms, haplotypes and in silico approach. Mol. Biol. Rep. 2020, 47, 7929–7940. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, Q.; Zhang, W.; Chen, F.; Zhao, T.; Lin, Y.; Li, J.; Liu, Y.; Liu, Y.; Shao, Y. The interleukin-27 -964A>G polymorphism enhances sepsis-induced inflammatory responses and confers susceptibility to the development of sepsis. Crit. Care 2018, 22, 248. [Google Scholar] [CrossRef] [Green Version]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okitsu, C.Y.; Hsieh, J.C.F.; Hsieh, C.-L. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 2010, 30, 2933–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghavami, A.; Fathpour, G.; Amirghofran, Z. Association of IL-27 rs153109 and rs17855750 Polymorphisms with Risk and Response to Therapy in Acute Lymphoblastic Leukemia. Pathol. Oncol. Res. 2018, 24, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.-C.; Li, C.-S.; Kim, K.M.; Yang, J.Y.; Zhang, Q.; Lee, Y.-C.; Yang, Y.-S.; Chung, H.-T. Identification of polymorphisms in human interleukin-27 and their association with asthma in a Korean population. J. Hum. Genet. 2007, 52, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- rs181206 (SNP)-Citations-Homo Sapiens-GRCh37 Archive Browser 100. Available online: http://grch37.ensembl.org/Homo_sapiens/Variation/Citations?db=core;g=ENSG00000197272;r=16:28510683-28518155;t=ENST00000356897;v=rs181206;vdb=variation;vf=309953932 (accessed on 3 August 2020).
- Si, F.; Wu, Y.; Wang, X.; Gao, F.; Yang, D.; Liu, R.; Yi, Q. The relationship between Interleukin-27 gene polymorphisms and Kawasaki disease in a population of Chinese children. Cardiol. Young 2018, 28, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Gholijani, N.; Daryabor, G.; Kalantar, K.; Yazdani, M.R.; Shenavandeh, S.; Zahed, M.; Jafarpour, Z.; Malekmakan, M.R.; Amirghofran, Z. Interleukin-27 gene variant rs153109 is associated with enhanced cytokine serum levels and susceptibility to Behçet’s disease in the Iranian population. Eur. Cytokine Netw. 2020, 31, 140–146. [Google Scholar] [CrossRef]
- Yamamoto-Furusho, J.K.; Posadas-Sánchez, R.; Alvarez-León, E.; Vargas-Alarcón, G. Protective role of Interleukin 27 (IL-27) gene polymorphisms in patients with ulcerative colitis. Immunol. Lett. 2016, 172, 79–83. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Anber, N.; El Sayed Zaki, M.; El Wassefy, M. Interleukin-27 levels and its Polymorphism in Hepatocellular carcinoma associated with Hepatitis C. Nov. Res. Microbiol. J. 2020, 4, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Mosayebian, A.; Hakemi, M.G.; Meshkat, R.; Ghasemi, R.; Ahmad, H.K.; Samadi, M. Association between interleukin-23 receptor R381Q gene polymorphism and asthma. Iran. J. Allergy Asthma Immunol. 2015, 14, 386–391. [Google Scholar]
- Mohammadi, S.A.; Mansouri, R.; Shahi, A.; Akhlaghi, M.; Dashti, N.; Aslani, S.; Mansouri, M.; Poursani, S.; Mahmoudi, M. IL27 gene single nucleotide polymorphisms confer susceptibility to rheumatoid arthritis in Iranian population. Meta Gene 2018, 18, 149–152. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. Quantitative trait loci analysis using the false discovery rate. Genetics 2005, 171, 783–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barac, I.S.; Văcăraș, V.; Cozma, A.; Procopciuc, L.M. Multiple sclerosis—Behind the immunity curtains. Ces. Slov. Neurol. Neurochir. 2020, 83, 368–374. [Google Scholar] [CrossRef]
- Volpe, E.; Battistini, L.; Borsellino, G. Advances in T Helper 17 Cell Biology: Pathogenic Role and Potential Therapy in Multiple Sclerosis. Mediat. Inflamm. 2015, 2015, 475158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rostami, A.; Ciric, B. Role of Th17 cells in the pathogenesis of CNS inflammatory demyelination. J. Neurol. Sci. 2013, 333, 76–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Langrish, C.L.; Mckenzie, B.; Joyce-Shaikh, B.; Stumhofer, J.S.; McClanahan, T.; Blumenschein, W.; Churakovsa, T.; Low, J.; Presta, L.; et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J. Clin. Investig. 2006, 116, 1317–1326. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Lovett-Racke, A.E.; Racke, M.K. Role of IL-12/IL-23 in the Pathogenesis of Multiple Sclerosis; Elsevier Inc.: Amsterdam, The Netherlands, 2011; ISBN 9780123849137. [Google Scholar]
- Abdollahi, E.; Tavasolian, F.; Momtazi-Borojeni, A.A.; Samadi, M.; Rafatpanah, H. Protective role of R381Q (rs11209026) polymorphism in IL-23R gene in immune-mediated diseases: A comprehensive review. J. Immunotoxicol. 2016, 13, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Yalçin, B.; Atakan, N.; Dogan, S. Association of interleukin-23 receptor gene polymorphism with Behçet disease. Clin. Exp. Dermatol. 2014, 39, 881–887. [Google Scholar] [CrossRef]
- Hazlett, J.; Stamp, L.K.; Merriman, T.; Highton, J.; Hessian, P.A. IL-23R rs11209026 polymorphism modulates IL-17A expression in patients with rheumatoid arthritis. Genes Immun. 2012, 13, 282–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarin, R.; Wu, X.; Abraham, C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc. Natl. Acad. Sci. USA 2011, 108, 9560–9565. [Google Scholar] [CrossRef] [Green Version]
- Di Meglio, P.; Villanova, F.; Napolitano, L.; Tosi, I.; Terranova Barberio, M.; Mak, R.K.; Nutland, S.; Smith, C.H.; Barker, J.N.W.N.; Todd, J.A.; et al. The IL23R A/Gln381 allele promotes IL-23 unresponsiveness in human memory T-helper 17 cells and impairs Th17 responses in psoriasis patients. J. Investig. Dermatol. 2013, 133, 2381–2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nossent, J.C.; Sagen-Johnsen, S.; Bakland, G. IL23R gene variants in relation to IL17A levels and clinical phenotype in patients with ankylosing spondylitis. Rheumatol. Adv. Pract. 2018, 2, rky006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filiz, B.; Yildirim, M.; Öztürk, K.H.; Şirin, F.B.; Çelik, S.; Erturan, İ.; Korkmaz, S.; Orhan, H. Evaluation of interleukin-23 receptor (Il-23r) gene polymorphisms and serum il-23 levels in patients with psoriasis. Turk. J. Med. Sci. 2019, 49, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wang, X.; Tan, G.; Liang, Z.; Zhang, Z.; Yu, H. The association between genetic polymorphisms of interleukin 23 receptor gene and the risk of rheumatoid arthritis: An updated meta-analysis. Clin. Immunol. 2020, 210, 108250. [Google Scholar] [CrossRef] [PubMed]
- Deveci, H.; Turk, A.C.; Ozmen, Z.C.; Demir, A.K.; Coskun, S.U.S. Biological and genetic evaluation of IL-23/IL-17 pathway in ankylosing spondylitis patients. Cent. Eur. J. Immunol. 2019, 44, 433–439. [Google Scholar] [CrossRef]
- Ghaffari, S.A.; Nemati, M.; Hajghani, H.; Ebrahimi, H.; Sheikhi, A.; Jafarzadeh, A. Circulating concentrations of interleukin (IL)-17 in patients with multiple sclerosis: Evaluation of the effects of gender, treatment, disease patterns and IL-23 receptor gene polymorphisms. Iran. J. Neurol. 2017, 16, 15–25. [Google Scholar] [PubMed]
- Nunez, C.; Dema, B.; Cenit, M.C.; Polanco, I.; Maluenda, C.; Arroyo, R.; de las Heras, V.; Bartolomé, M.; de la Concha, E.G.; Urcelay, E.; et al. IL23R: A susceptibility locus for celiac disease and multiple sclerosis? Genes Immun. 2008, 9, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Choi, D.; Lim, L.L.; Allada, G.; Smith, J.R.; Austin, C.R.; Doyle, T.M.; Goodwin, K.A.; Rosenbaum, J.T.; Martin, T.M. Association of interleukin 23 receptor gene with sarcoidosis. Dis. Markers 2011, 31, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, M.; Wandtke, T. Interleukin-27: Biological Properties and Clinical Application, 1st ed.; SpringerBriefs in Immunology; Springer: Cham, Denmark, 2016; ISBN 978-3-319-39662-0. [Google Scholar]
- Pflanz, S.; Timans, J.C.; Cheung, J.; Rosales, R.; Kanzler, H.; Gilbert, J.; Hibbert, L.; Churakova, T.; Travis, M.; Vaisberg, E.; et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+T cells. Immunity 2002, 16, 779–790. [Google Scholar] [CrossRef] [Green Version]
- Hirahara, K.; Ghoreschi, K.; Yang, X.-P.; Takahashi, H.; Laurence, A.; Vahedi, G.; Sciumè, G.; Hall, A.O.; Dupont, C.D.; Francisco, L.M.; et al. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity 2012, 36, 1017–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasanthakumar, A.; Kallies, A. IL-27 paves different roads to Tr1. Eur. J. Immunol. 2013, 43, 882–885. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Chen, P.; Zhou, B.; Zhang, P.; Wang, Y.; Song, Y.; Zhang, L. Association between polymorphisms in IL27 gene and renal cell carcinoma. Biomarkers 2015, 20, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.; Ye, L.; Wang, O.; Huang, G.; Yang, F.; Liu, Y.; Dong, S. IL-27 rs153109 polymorphism increases the risk of colorectal cancer in Chinese Han population. Onco Targets Ther. 2015, 8, 1493. [Google Scholar] [CrossRef] [Green Version]
- Moazeni-Roodi, A.; Hashemi, M.; Ghavami, S. Association between IL-27 Gene Polymorphisms and Cancer Susceptibility in Asian Population: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2020, 21, 2507–2515. [Google Scholar] [CrossRef]
- Dehghanzadeh, R.; Babaloo, Z.; Sakhinia, E.; Khabazi, A.; Shanehbandi, D.; Sadigh-Eteghad, S.; Gharibi, T. IL-27 Gene Polymorphisms in Iranian Patients with Behcet’s Disease. Clin. Lab. 2016, 62, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Paradowska-Gorycka, A.; Raszkiewicz, B.; Jurkowska, M.; Felis-Giemza, A.; Romanowska-Prochnicka, K.; Mańczak, M.; Olesinska, M. Association of single nucleotide polymorphisms in the IL27 gene with rheumatoid arthritis. Scand. J. Immunol. 2014, 80, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, K.; Wang, Z.; Zhang, J.; Chen, T.; Jin, L. Functional variant in the promoter region of IL-27 alters gene transcription and confers a risk for ulcerative colitis in northern Chinese Han. Hum. Immunol. 2017, 78, 287–293. [Google Scholar] [CrossRef]
- Morita, Y.; Masters, E.A.; Schwarz, E.M.; Muthukrishnan, G. Interleukin-27 and Its Diverse Effects on Bacterial Infections. Front. Immunol. 2021, 12, 1752. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-S.; Zhang, Q.; Lee, K.-J.; Cho, S.-W.; Lee, K.-M.; Hahm, K.-B.; Choi, S.-C.; Yun, K.-J.; Chung, H.-T.; Chae, S.-C. Interleukin-27 polymorphisms are associated with inflammatory bowel diseases in a Korean population. J. Gastroenterol. Hepatol. 2009, 24, 1692–1696. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, Y.; Xue, F.; Xu, J.; Fang, Z. Interleukin-27 rs153109 polymorphism and the risk for immune thrombocytopenia. Autoimmunity 2013, 46, 509–512. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, B.; Mu, K.; Zhang, J.; Yang, Y.; Yao, W.; Li, S.; Zhang, J. Association of single-nucleotide polymorphisms in the IL27 gene with autoimmune thyroid diseases. Endocr. Connect. 2019, 8, 173–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Yuan, X.D.; Hu, D.; Ke, X.; Wang, X.Q.; Hu, G.H.; Hong, S.L.; Kang, H.Y. Association between Interleukin-27 gene polymorphisms and susceptibility to allergic rhinitis. Hum. Immunol. 2014, 75, 991–995. [Google Scholar] [CrossRef]
- Vargas-Alarcón, G.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.; Fragoso, J.M.; Posadas-Romero, C.; López-Bautista, F.; Vázquez-Vázquez, C.; Posadas-Sánchez, R. Interleukin 27 polymorphisms, their association with insulin resistance and their contribution to subclinical atherosclerosis. The GEA Mexican study. Cytokine 2019, 114, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Luo, S.-D.; Zhang, T.; Shi, F.; Wang, C.-F.; Chen, X.; Wei, Y.-X.; Qin, L.; Wei, J.; Luo, X.; et al. Association of interleukin-27 gene polymorphisms with susceptibility to HIV infection and disease progression. J. Cell. Mol. Med. 2019, 23, 2410–2418. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Li, Y.; Yao, Y.; Li, H.; Liang, H.; Xin, M.; Wang, L.; Zhao, L.; Lin, J.; Liu, S. Polymorphisms of the IL27 gene in a Chinese Han population complicated with pre-eclampsia. Sci. Rep. 2016, 6, 23029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barac, I.S.; Vacaras, V.; Cozma, A.; Valeanu, M.; Decea, N.; Muresanu, D.F.; Procopciuc, L.M. Il27 t4730c polymorphism and serology in multiple sclerosis: A pilot study. In Vivo 2021, 35, 2845–2853. [Google Scholar] [CrossRef]
- Huang, N.; Liu, L.; Wang, X.Z.; Liu, D.; Yin, S.Y.; Yang, X.D. Association of interleukin (IL)-12 and IL-27 gene polymorphisms with chronic obstructive pulmonary disease in a Chinese population. DNA Cell Biol. 2008, 27, 527–531. [Google Scholar] [CrossRef] [PubMed]
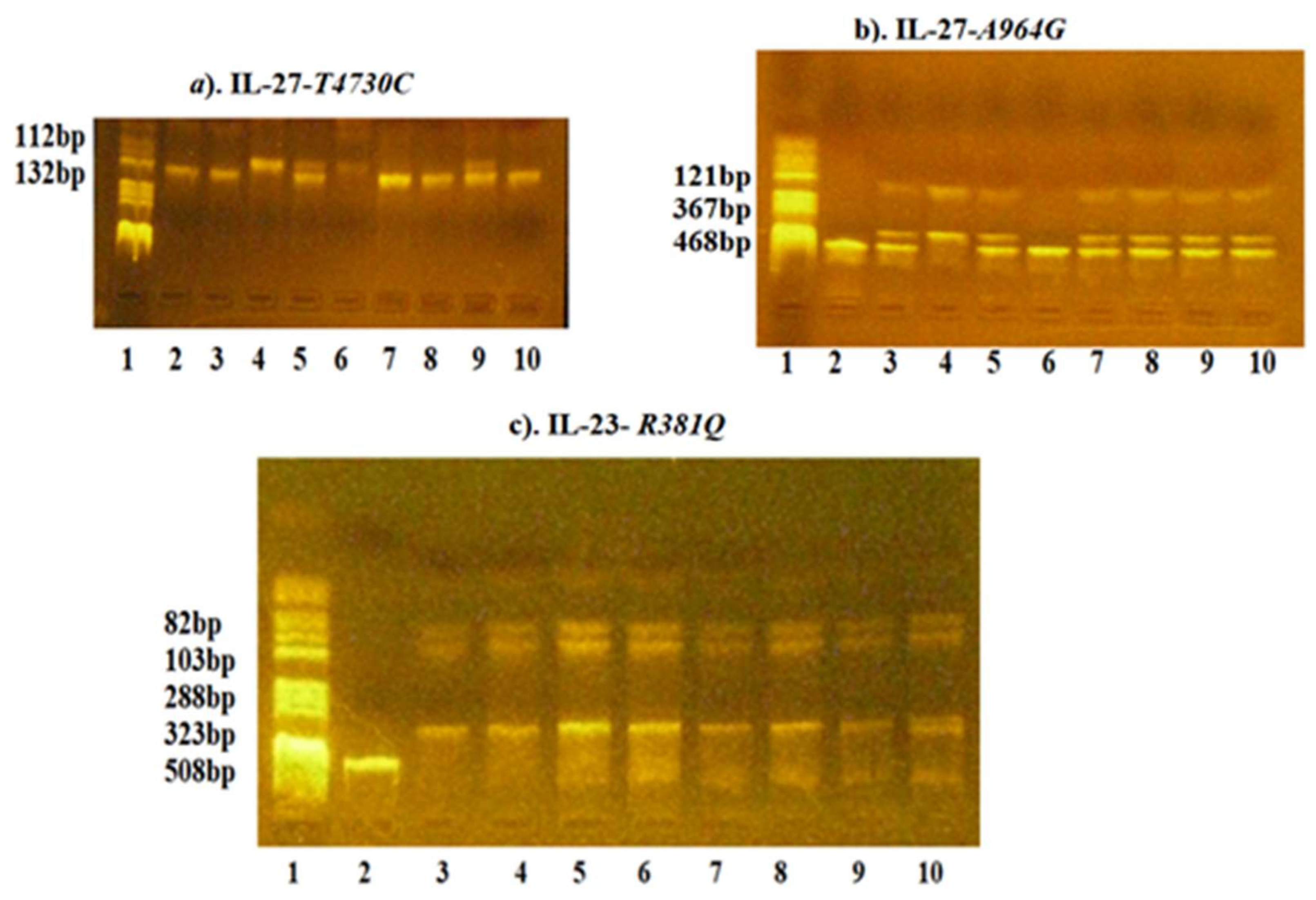
| Genetic Variations | Primers | Tanneal | PCR Fragments | Restriction Enzymes | Fragments |
|---|---|---|---|---|---|
| IL-27-T4730C (Leu119Pro) (rs 181208) | FW:5′- GCTTCAGCCCTTCCATGCCC-3′ RW:5′-TCTACCTGGAAGCGGAGGTGCC-3′ | 67 °C | 132 bp | FuaI | T4730 allele: 132 bp C4730 allele: 112, 20 bp |
| IL-27-A964G (rs 153109) | FW:5′-GGCTGTGCTGGAAGGGAGAC-3′ RW: 5-ATATCTGGGACCAGGGTTAGG-3′ | 62 °C | 468 bp | XhoI | A964 allele: 468 bp G964 allele: 347, 121 bp |
| IL-23-G1142A (R381Q) (rs 11209026) | FW:5′-CTTTTCTGGCAGGGTCATTTTG-3′ RW:5′-AAGTTGTTTCCTGGGGTAGTTGTG-3′ | 60 °C | 508 bp | Hpy188i | G1142 allele: 288, 103, 82, 35 bp A1142 allele: 323, 35 bp |
| Category Data | Variables | Controls (n1 = 95) | MS Patients (n2 = 157) | p-Value | |
|---|---|---|---|---|---|
| Demographic | Age, years (a) | 27 (25,32) | 42 (31,50) | <0.001 * | |
| Sex (b) | |||||
| Male | 34 (35.8) | 51 (32.5) | 0.591 | ||
| Female | 61 (64.2) | 106 (67.5) | |||
| Residence (b) | |||||
| Urban area | 95 (100.0) | 122 (77.7) | <0.001 * | ||
| Rural area | 0 (0.0) | 35 (22.3) | |||
| Lifestyle | Smoking (b) | 20 (21.1) | 52 (33.1) | 0.040 * | |
| Alcohol consumption (b) | 59 (62.1) | 35 (22.4) | <0.001 * | ||
| Clinical | Age at diagnosis, years (c) | NA | 30 (24, 40) | ||
| Duration of the disease (b) | |||||
| <5 years | NA | 62 (39.5) | |||
| 5–10 years | NA | 27 (17.2) | |||
| >5 years | NA | 68 (43.3) | |||
| Form of MS (b) | |||||
| CIS | NA | 26 (16.6) | |||
| RR | NA | 109 (69.4) | |||
| SP | NA | 22 (14.0) | |||
| EDSS (c) | NA | 2.0 (1.0, 3.0) | |||
| Disease treatment (a) | NA | 109 (69.4) |
| SNPs | Genetic Model | Genotypes | Controls (a) (n1 = 95) | MS Patients (a) (n2 = 157) | OR [95% CI] | p-Value Unadjusted | OR [95% CI] (b) | p-Value Adjusted (b) | Q-Value (c) |
|---|---|---|---|---|---|---|---|---|---|
| IL-27-T4730C | Codominant | TT | 76 (80.0) | 86 (54.8) | 1.00 | 0.0002 * | 1.00 | 0.00004 * | 0.00018 * |
| TC | 15 (15.8) | 52 (33.1) | 3.06 [1.60, 5.88] | 4.16 [2.04, 8.48] | |||||
| CC | 4 (4.2) | 19 (12.1) | 4.20 [1.37, 12.88] | 3.87 [1.17, 12.81] | |||||
| Dominant | TT | 76 (80.0) | 86 (54.8) | 1.00 | 0.00003 * | 1.00 | 0.000007 * | 0.000063 * | |
| TC + CC | 19 (20.0) | 71 (45.2) | 3.30 [1.83, 5.97] | 4.09 [2.14, 7.83] | |||||
| Recessive | TT + TC | 91 (95.8) | 138 (87.9) | 1.00 | 0.026 * | 1.00 | 0.083 | 0.1067 | |
| CC | 4 (4.2) | 19 (12.1) | 3.13 [1.03, 9.51] | 2.64 [0.82, 8.51] | |||||
| Overdominant | TT + CC | 80 (84.2) | 105 (66.9) | 1.00 | 0.00019 * | 1.00 | 0.00015 * | 0.00045 * | |
| TC | 15 (15.8) | 52(33.1) | 3.13 [1.03, 9.51] | 3.64 [1.80, 7.36] | |||||
| IL-27-A964G | Codominant | AA | 37 (38.9) | 39 (24.8) | 1.00 | 0.0428 * | 1.00 | 0.0727 | 0.1067 |
| AG | 44 (46.3) | 82 (52.2) | 1.77 [0.99, 3.16] | 1.77 [0.94, 3.33] | |||||
| GG | 14 (14.7) | 36 (22.9) | 2.44 [1.14, 5.24] | 2.43 [1.06, 5.55] | |||||
| Dominant | AA | 37 (38.9) | 39 (24.8) | 1.00 | 0.0189 * | 1.00 | 0.0324 * | 0.05832 # | |
| AG + GG | 58 (61.1) | 118 (75.2) | 1.93 [1.12, 3.34] | 1.93 [1.05, 3.51] | |||||
| Recessive | AA + AG | 81 (85.3) | 121 (77.1) | 1.00 | 0.1082 | 1.00 | 0.1425 | 0.16031 | |
| GG | 14 (14.7) | 36 (22.9) | 1.72 [0.87, 3.39] | 1.71 [0.82, 3.55] | |||||
| Overdominant | AA + GG | 51 (53.7) | 75 (47.8) | 1.00 | 0.3627 | 1.00 | 0.4075 | 0.4075 | |
| AG | 44 (46.3) | 82(52.2) | 1.27 [0.76, 2.11] | 1.26 [0.73, 2.20] | |||||
| IL-23-R381Q | Codominant | GG | 85 (89.5) | 153 (97.5) | 1.00 | 0.0084 * | 1.00 | 0.0276 * | 0.05832 # |
| AG | 10 (10.5) | 4 (2.5) | 0.22 [0.07, 0.73] | 0.26 [0.08, 0.92] | |||||
| AA | 0 (0.0) | 0 (0.0) | ─ | ─ |
| Gene Polymorphisms | Genetic Model | IL-27-A964G | IL-27-T4730C | IL-23-R381Q |
|---|---|---|---|---|
| IL-27-A964G | Codominant | 0.0423/0.069 | 0.294/0.195 | 0.106/0.267 |
| Dominant | 0.018/0.029 | 0.063/0.093 | 0.089/0.171 | |
| Recessive | 0.115/0.155 | 0.584/0.575 | NA | |
| IL-27-T4730C | Codominant | 0.0002/<0.001 | 0.536/0.269 | |
| Dominant | <0.0001/<0.0001 | 0.511/0.282 | ||
| Recessive | 0.035/0.099 | NA | ||
| IL-23-R381Q | Codominant | 0.007/0.024 | ||
| Dominant | 0.007/0.024 | |||
| Recessive | NA |
| Haplotypes | Total Relative Frequency | Haplotype Frequencies in the Control Group | Haplotype Frequencies in MS Patients | Hap-Score (a) | p-Value (b) | Permutation p-Value (c) | OR (d) [95% CI] | OR (e) [95% CI] |
|---|---|---|---|---|---|---|---|---|
| A—T | 0.5074 | 0.5809 | 0.4631 | −2.715 | 0.0066 * | 0.0060 * | 1.00 [Reference] | 1.00 [Reference] |
| G—T | 0.2684 | 0.2980 | 0.2503 | −0.998 | 0.3174 | 0.3187 | 1.14 [0.73, 1.76] | 1.10 [0.71, 1.71] |
| A—C | 0.0442 | 0.0401 | 0.0465 | 0.709 | 0.4782 | 0.4748 | 1.32 [0.47, 3.73] | 1.37 [0.48, 3.87] |
| G—C | 0.1799 | 0.0809 | 0.2401 | 4.02 | 0.00006 * | 0.00005 * | 3.20 [1.74, 5.88] | 3.18 [1.74, 5.81] |
| Variables | IL-27-A964G | IL-27-T4730C | IL-23-R381Q | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AA (n1 = 39) | AG/GG (n2 = 118) | p-Value | TT (n1 = 86) | TC/CC (n2 = 71) | p-Value | GG (n1 = 153) | AG (n2 = 4) | p-Value | |
| Age, years (a) | 42.00 ± 12.58 | 40.97 ± 10.76 | 0.619 | 42.19 ± 11.88 | 40.06 ± 10.29 | 0.237 | 41.20 ± 10.98 | 42.00 ± 20.38 | 0.888 |
| Age at diagnosis, years (b) | 30 (23.5, 39.5) | 30 (26,40) | 0.642 | 29.5 (24.25, 39.75) | 30 (25,40) | 0.951 | 30 (25,40) | 24.50 (22.25, 30) | 0.308 |
| Sex (male) (c) | 15 (38.46) | 36 (30.51) | 0.358 | 28 (32.56) | 23 (32.39) | 0.983 | 50 (32.68) | 1 (25.00) | 1.000 |
| Residence (urban) (c) | 28 (71.79) | 94 (79.66) | 0.306 | 68 (79.07) | 54 (76.06) | 0.652 | 118 (77.12) | 4 (100.00) | 0.576 |
| Smoking (c) | 11 (28.21) | 41 (34.75) | 0.452 | 28 (32.56) | 24 (33.80) | 0.869 | 50 (32.68) | 2 (50.00) | 0.600 |
| Alcohol consumption (c) | 8 (20.51) | 27 (23.08) | 0.739 | 19 (22.35) | 16 (22.54) | 0.978 | 33 (21.71) | 2 (50.00) | 0.218 |
| Duration of the disease (c) | 0.507 | 0.507 | 0.252 | 0.291 | |||||
| <5 years | 13 (33.33) | 49 (41.53) | 32 (37.21) | 30 (42.25) | 61 (39.87) | 1 (25.00) | |||
| 5–10 years | 6 (15.38) | 21 (17.80) | 12 (13.95) | 15 (21.13) | 25 (16.34) | 2 (50.00) | |||
| >10 years | 20 (51.28) | 48 (40.68) | 42 (48.84) | 26 (36.62) | 67 (43.79) | 1 (25.00) | |||
| Personal history of autoimmune diseases (c) | 1 (2.56) | 4 (3.39) | 1.000 | 3 (3.49) | 4 (2.82) | 1.000 | 4 (2.61) | 1 (25.00) | 0.123 |
| Form of MS (c) | 0.091 | 0.259 | 0.772 | ||||||
| CIS | 8 (20.51) | 18 (15.25) | 17 (19.77) | 9 (12.68) | 25 (16.34) | 1 (25.00) | |||
| RR | 22 (56.41) | 87 (73.73) | 55 (63.95) | 54 (76.06) | 106 (69.28) | 3 (75.00) | |||
| SP | 9 (23.08) | 13 (11.02) | 14 (16.28) | 8 (11.27) | 22 (14.38) | 0 (0.00) | |||
| EDSS Score at admission (b) | 2.0 (1.0, 3.5) | 2.0 (1.0, 3.0) | 0.399 | 2.0 (1.0, 3.0) | 2.0 (1.0, 3.5) | 0.793 | 2.0 (1.0, 3.5) | 1.5 (1.4, 1.8) | 0.577 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barac, I.S.; Iancu, M.; Văcăraș, V.; Cozma, A.; Negrean, V.; Sâmpelean, D.; Mureșanu, D.F.; Procopciuc, L.M. Potential Contribution of IL-27 and IL-23 Gene Polymorphisms to Multiple Sclerosis Susceptibility: An Association Analysis at Genotype and Haplotype Level. J. Clin. Med. 2022, 11, 37. https://doi.org/10.3390/jcm11010037
Barac IS, Iancu M, Văcăraș V, Cozma A, Negrean V, Sâmpelean D, Mureșanu DF, Procopciuc LM. Potential Contribution of IL-27 and IL-23 Gene Polymorphisms to Multiple Sclerosis Susceptibility: An Association Analysis at Genotype and Haplotype Level. Journal of Clinical Medicine. 2022; 11(1):37. https://doi.org/10.3390/jcm11010037
Chicago/Turabian StyleBarac, Ioana S., Mihaela Iancu, Vitalie Văcăraș, Angela Cozma, Vasile Negrean, Dorel Sâmpelean, Dafin F. Mureșanu, and Lucia M. Procopciuc. 2022. "Potential Contribution of IL-27 and IL-23 Gene Polymorphisms to Multiple Sclerosis Susceptibility: An Association Analysis at Genotype and Haplotype Level" Journal of Clinical Medicine 11, no. 1: 37. https://doi.org/10.3390/jcm11010037
APA StyleBarac, I. S., Iancu, M., Văcăraș, V., Cozma, A., Negrean, V., Sâmpelean, D., Mureșanu, D. F., & Procopciuc, L. M. (2022). Potential Contribution of IL-27 and IL-23 Gene Polymorphisms to Multiple Sclerosis Susceptibility: An Association Analysis at Genotype and Haplotype Level. Journal of Clinical Medicine, 11(1), 37. https://doi.org/10.3390/jcm11010037









