Proteomic Insight into the Role of Exosomes in Proliferative Vitreoretinopathy Development
Abstract
:1. Introduction
2. Material and Methods
2.1. Patients
2.2. Vitreous Humor (VH) Preparation
2.3. Exosome Isolation and Characterization
2.4. Proteomic Analysis
3. Data and Bioinformatics Analysis
4. Results
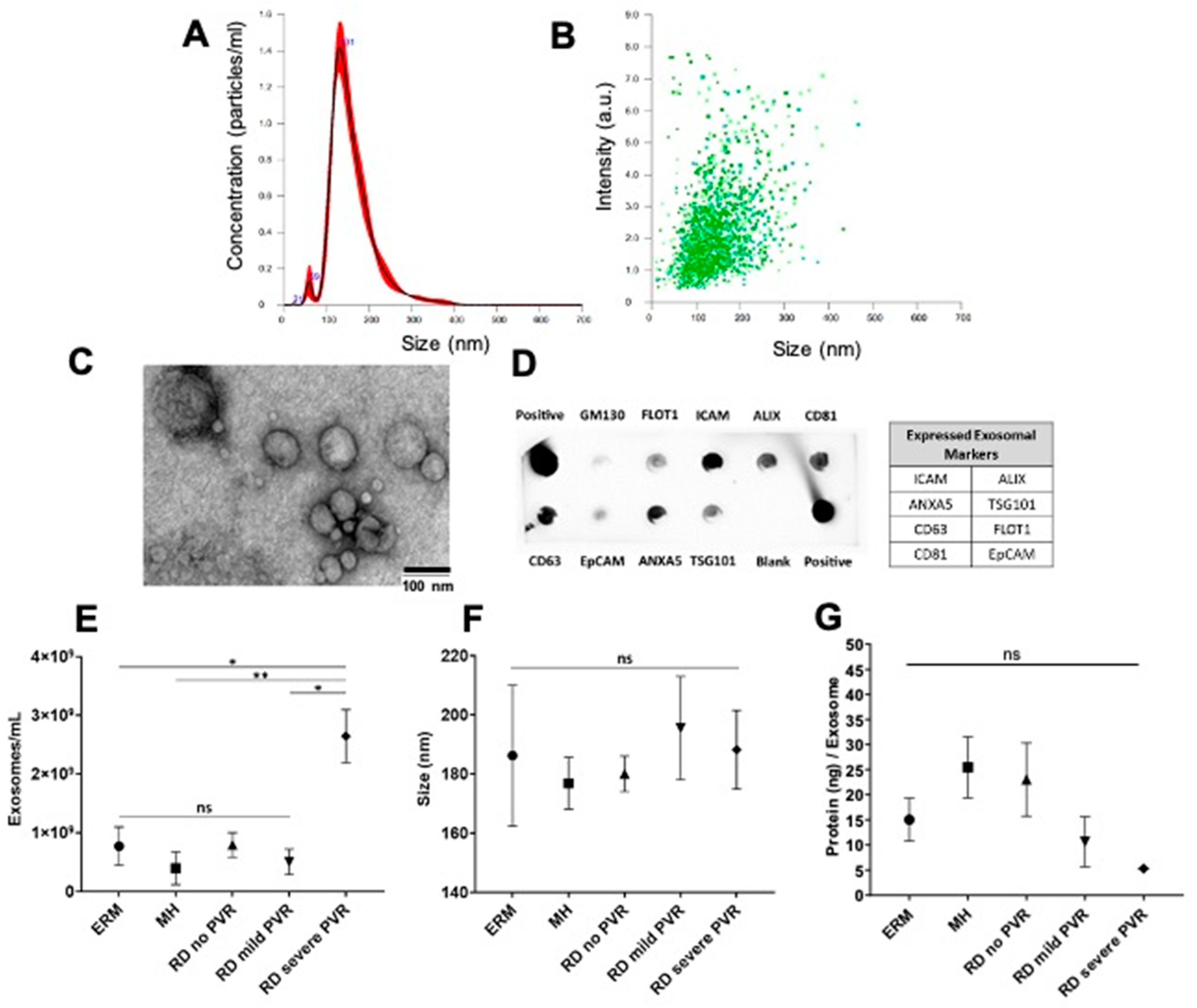
4.1. Characterization of Exosomes Isolated from Vitreous Humor (VH)
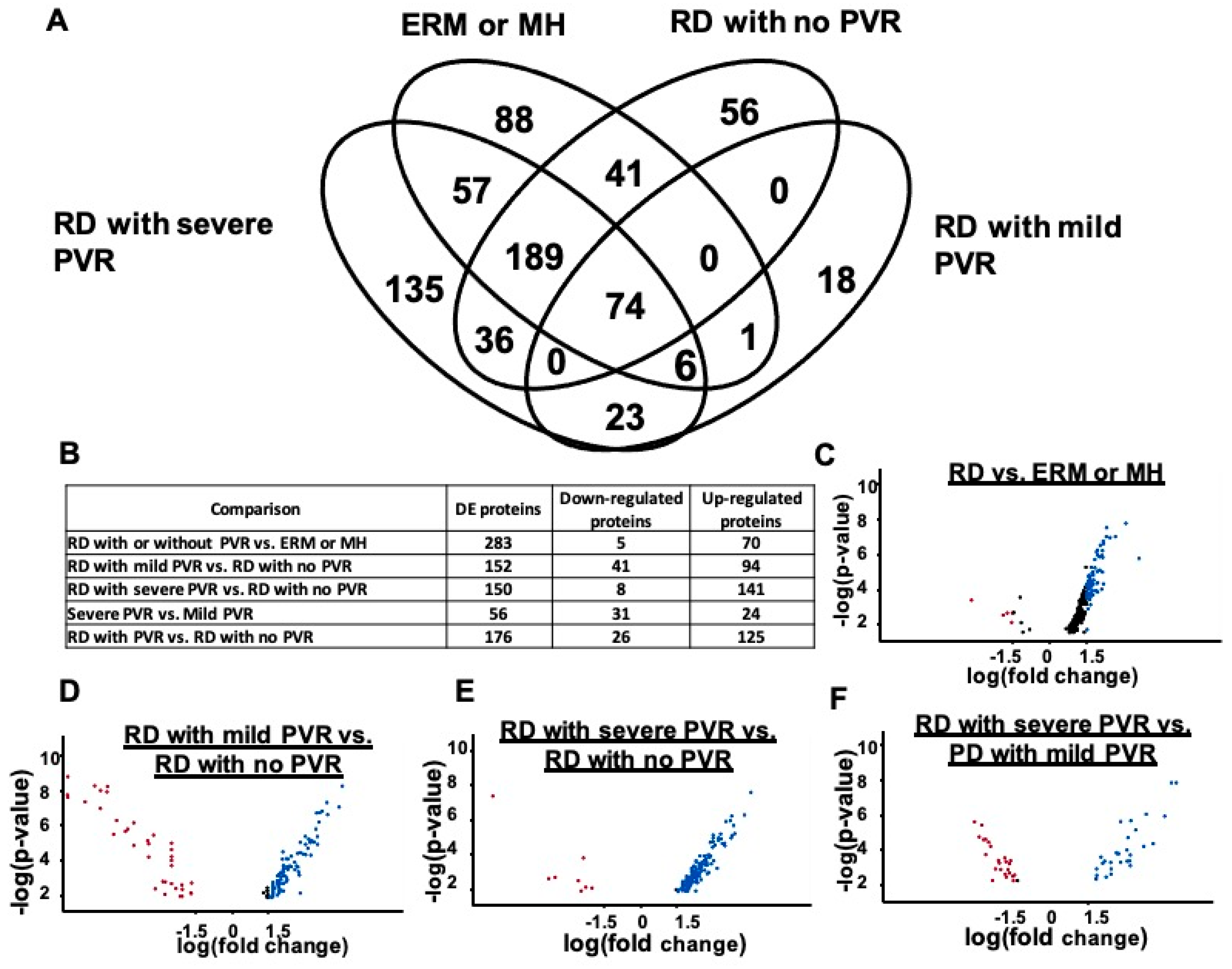
4.2. Proteins Detected in Exosomes with PVR and Differential Protein Expression
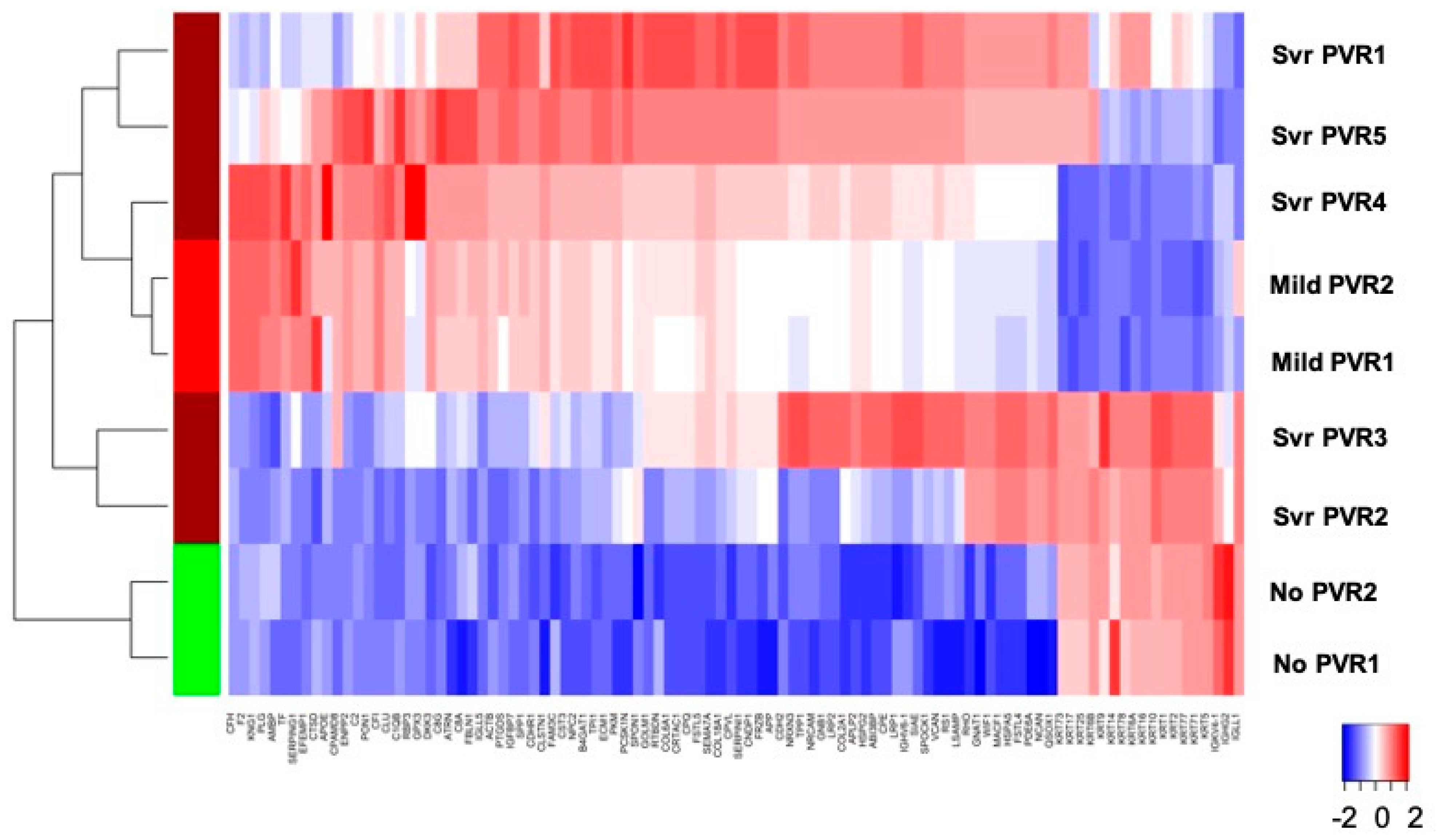
4.3. Stage-Specific Protein Expression
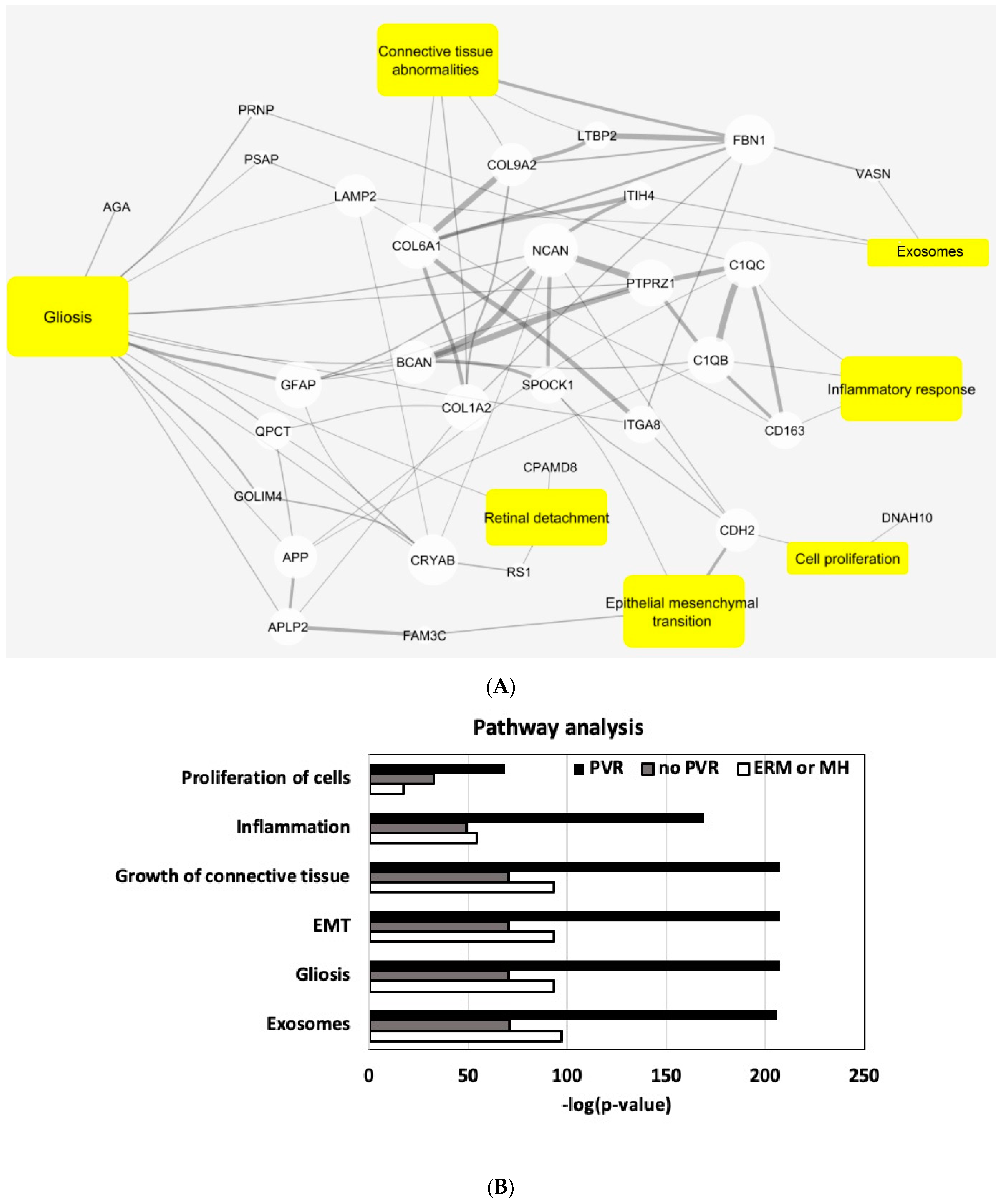
4.4. Pathway Analysis
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pastor, J.C.; de la Rua, E.R.; Marti, F. Proliferative vitreoretinopathy: Risk factors and pathobiology. Prog. Retin. Eye Res. 2002, 21, 127–144. [Google Scholar] [CrossRef]
- Lee, S. Pathophysiology of Ocular Trauma. In Ryan’s Retina, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2017; Chapter 102; pp. 1685–1875. [Google Scholar]
- Kuriyan, A.E.; Albini, T.A.; Townsend, J.H.; Rodriguez, M.; Pandya, H.K.; Leonard, R.E., 2nd; Parrott, M.B.; Rosenfeld, P.J.; Flynn, H.W., Jr.; Goldberg, J.L. Vision Loss after Intravitreal Injection of Autologous “Stem Cells” for AMD. N. Engl. J. Med. 2017, 376, 1047–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campochiaro, P.A. Pathogenic Mechanisms in Proliferative Vitreoretinopathy. Arch. Ophthalmol. 1997, 115, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wiedemann, P.; Heimann, K. Proliferative vitreoretinopathy—Is it anything more than wound healing at the wrong place? Int. Ophthalmol. 1990, 14, 105–117. [Google Scholar] [CrossRef]
- Mudhar, H.S. A brief review of the histopathology of proliferative vitreoretinopathy (PVR). Eye 2019, 34, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M. Fibrosis and diseases of the eye. J. Clin. Investig. 2007, 117, 576–586. [Google Scholar] [CrossRef]
- Tamiya, S.; Kaplan, H.J. Role of epithelial–mesenchymal transition in proliferative vitreoretinopathy. Exp. Eye Res. 2016, 142, 26–31. [Google Scholar] [CrossRef]
- Hinz, B. Myofibroblasts. Exp. Eye Res. 2016, 142, 56–70. [Google Scholar] [CrossRef]
- Shu, D.Y.; Lovicu, F.J. Myofibroblast transdifferentiation: The dark force in ocular wound healing and fibrosis. Prog. Retin. Eye Res. 2017, 60, 44–65. [Google Scholar] [CrossRef]
- Wiedemann, P.; Hilgers, R.-D.; Bauer, P.; Heimann, K. Adjunctive daunorubicin in the treatment of proliferative vitreoretinopathy: Results of a multicenter clinical trial. Am. J. Ophthalmol. 1998, 126, 550–559. [Google Scholar] [CrossRef]
- Asaria, R.H.Y.; Kon, C.H.; Bunce, C.; Charteris, D.G.; Wong, D.; Khaw, P.T.; Aylward, G.W. Adjuvant 5-fluorouracil and heparin prevents proliferative vitreoretinopathy: Results from a randomized, double-blind, controlled clinical trial. Ophthalmology 2001, 108, 1179–1183. [Google Scholar] [CrossRef]
- Banerjee, P.J.; Quartilho, A.; Bunce, C.; Xing, W.; Zvobgo, T.M.; Harris, N.; Charteris, D.G. Slow-Release Dexamethasone in Proliferative Vitreoretinopathy: A Prospective, Randomized Controlled Clinical Trial. Ophthalmology 2017, 124, 757–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilton, G.; Machemer, R.; Michels, R.; Okun, E.; Schepens, C.; Schwartz, A. The Classification of Retinal Detachment with Proliferative Vitreoretinopathy. Ophthalmology 1983, 90, 121–125. [Google Scholar] [CrossRef]
- Lee, S.Y.; Surbeck, J.W.; Drake, M.; Saunders, A.; Jin, H.D.; Shah, V.A.; Rajala, R.V. Increased Glial Fibrillary Acid Protein and Vimentin in Vitreous Fluid as a Biomarker for Proliferative Vitreoretinopathy. Investig. Opthalmol. Vis. Sci. 2020, 61, 22. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowskawieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, A.-K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Wang, X.; Yang, H.; Li, J.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef] [Green Version]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2014, 356, 339–346. [Google Scholar] [CrossRef]
- Klingeborn, M.; Dismuke, W.M.; Rickman, C.B.; Stamer, W.D. Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye Res. 2017, 59, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; Barbagallo, D.; et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol. Ther. 2015, 16, 1387–1396. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Weber, S.R.; Lease, J.; Russo, M.; Siedlecki, C.A.; Xu, L.C.; Chen, H.; Wang, W.; Ford, M.; Simó, R.; et al. Liquid Biopsy of Vitreous Reveals an Abundant Vesicle Population Consistent With the Size and Morphology of Exosomes. Transl. Vis. Sci. Technol. 2018, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Galardi, A.; Colletti, M.; Lavarello, C.; Di Paolo, V.; Mascio, P.; Russo, I.; Cozza, R.; Romanzo, A.; Valente, P.; De Vito, R.; et al. Proteomic Profiling of Retinoblastoma-Derived Exosomes Reveals Potential Biomarkers of Vitreous Seeding. Cancers 2020, 12, 1555. [Google Scholar] [CrossRef]
- Ahmad, M.T.; Zhang, P.; Dufresne, C.; Ferrucci, L.; Semba, R.D. The Human Eye Proteome Project: Updates on an Emerging Proteome. Proteomics 2018, 18, e1700394. [Google Scholar] [CrossRef]
- Wren, J.D.; Bekeredjian, R.; Stewart, J.A.; Shohet, R.V.; Garner, H.R. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics 2004, 20, 389–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Qian, W.J.; Gritsenko, M.A.; Camp, D.G., 2nd; Monroe, M.E.; Moore, R.J.; Smith, R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005, 4, 2070–2080. [Google Scholar] [CrossRef] [Green Version]
- Amador, P.B.; Royo, F.; González, E.; Vancells, J.C.; Palomo-Diez, L.; Borras, F.E.; Falcon-Perez, J.M. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J. Proteom. 2012, 75, 3574–3584. [Google Scholar] [CrossRef]
- Principe, S.; Jones, E.E.; Kim, Y.; Sinha, A.; Nyalwidhe, J.O.; Brooks, J.; Semmes, O.J.; Troyer, D.A.; Lance, R.S.; Kislinger, T.; et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics 2013, 13, 1667–1671. [Google Scholar] [CrossRef] [Green Version]
- Prunotto, M.; Farina, A.; Lane, L.; Pernin, A.; Schifferli, J.; Hochstrasser, D.F.; Lescuyer, P.; Moll, S. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J. Proteom. 2013, 82, 193–229. [Google Scholar] [CrossRef]
- Gonzalez-Begne, M.; Lu, B.; Han, X.; Hagen, F.K.; Hand, A.R.; Melvin, J.E.; Yates, I.J.R. Proteomic Analysis of Human Parotid Gland Exosomes by Multidimensional Protein Identification Technology (MudPIT). J. Proteome Res. 2009, 8, 1304–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J. Am. Soc. Nephrol. 2008, 20, 363–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Peng, R.; Chen, H.; Cui, C.; Ba, J. Elucidation of the Pathogenic Mechanism of Rhegmatogenous Retinal Detachment with Proliferative Vitreoretinopathy by Proteomic Analysis. Investig. Opthalmology Vis. Sci. 2012, 53, 8146–8153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Liu, F.; Cui, S.-J.; Liu, Y.; Song, Z.-Y.; Cao, H.; Chen, F.-E.; Wang, W.-J.; Sun, T.; Wang, F. Vitreous proteomic analysis of proliferative vitreoretinopathy. Proteomics 2008, 8, 3667–3678. [Google Scholar] [CrossRef] [PubMed]
- Roybal, C.N.; Velez, G.; Toral, M.A.; Tsang, S.H.; Bassuk, A.G.; Mahajan, V.B. Personalized Proteomics in Proliferative Vitreoretinopathy Implicate Hematopoietic Cell Recruitment and mTOR as a Therapeutic Target. Am. J. Ophthalmol. 2018, 186, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.-B.; Chen, X.; Timothy, N.; Aiello, L.P.; Feener, E.P. Characterization of the Vitreous Proteome in Diabetes without Diabetic Retinopathy and Diabetes with Proliferative Diabetic Retinopathy. J. Proteome Res. 2008, 7, 2516–2525. [Google Scholar] [CrossRef]
- Velez, G.; Yang, J.; Li, A.S.; Tsang, S.H.; Bassuk, A.; Mahajan, V.B. Proteomic insight into the pathogenesis of CAPN5-vitreoretinopathy. Sci. Rep. 2019, 9, 7608. [Google Scholar] [CrossRef]
- Zhang, Z.; Mugisha, A.; Fransisca, S.; Liu, Q.; Xie, P.; Hu, Z. Emerging Role of Exosomes in Retinal Diseases. Front. Cell Dev. Biol. 2021, 9, 643680. [Google Scholar] [CrossRef]
- Morris, D.R.; Bounds, S.E.; Liu, H.; Ding, W.-Q.; Chen, Y.; Liu, Y.; Cai, J. Exosomal MiRNA Transfer between Retinal Microglia and RPE. Int. J. Mol. Sci. 2020, 21, 3541. [Google Scholar] [CrossRef]
- Obermann, J.; Priglinger, C.S.; Merl-Pham, J.; Geerlof, A.; Priglinger, S.; Götz, M.; Hauck, S.M. Proteome-wide Identification of Glycosylation-dependent Interactors of Galectin-1 and Galectin-3 on Mesenchymal Retinal Pigment Epithelial (RPE) Cells. Mol. Cell. Proteom. 2017, 16, 1528–1546. [Google Scholar] [CrossRef] [Green Version]
- Priglinger, C.S.; Obermann, J.; Szober, C.M.; Merl-Pham, J.; Ohmayer, U.; Behler, J.; Gruhn, F.; Kreutzer, T.C.; Wertheimer, C.; Geerlof, A.; et al. Epithelial-to-Mesenchymal Transition of RPE Cells In Vitro Confers Increased β1,6-N-Glycosylation and Increased Susceptibility to Galectin-3 Binding. PLoS ONE 2016, 11, e0146887. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nair, G.K.G.; Pollalis, D.; Wren, J.D.; Georgescu, C.; Sjoelund, V.; Lee, S.Y. Proteomic Insight into the Role of Exosomes in Proliferative Vitreoretinopathy Development. J. Clin. Med. 2022, 11, 2716. https://doi.org/10.3390/jcm11102716
Nair GKG, Pollalis D, Wren JD, Georgescu C, Sjoelund V, Lee SY. Proteomic Insight into the Role of Exosomes in Proliferative Vitreoretinopathy Development. Journal of Clinical Medicine. 2022; 11(10):2716. https://doi.org/10.3390/jcm11102716
Chicago/Turabian StyleNair, Gopa Kumar Gopinadhan, Dimitrios Pollalis, Jonathan D. Wren, Constantin Georgescu, Virginie Sjoelund, and Sun Young Lee. 2022. "Proteomic Insight into the Role of Exosomes in Proliferative Vitreoretinopathy Development" Journal of Clinical Medicine 11, no. 10: 2716. https://doi.org/10.3390/jcm11102716





