A Novel Approach for the Shape Characterisation of Non-Melanoma Skin Lesions Using Elliptic Fourier Analyses and Clinical Images
Abstract
:1. Introduction
2. Materials and Methods
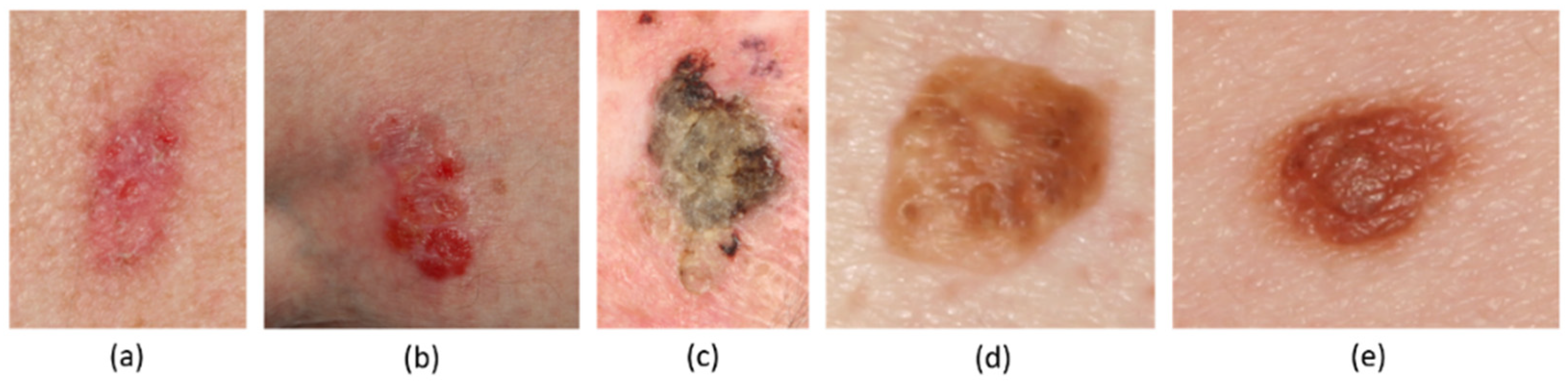
2.1. Image Dataset
2.2. Definition of Lesion Boundaries
2.3. Elliptic Fourier Analyses
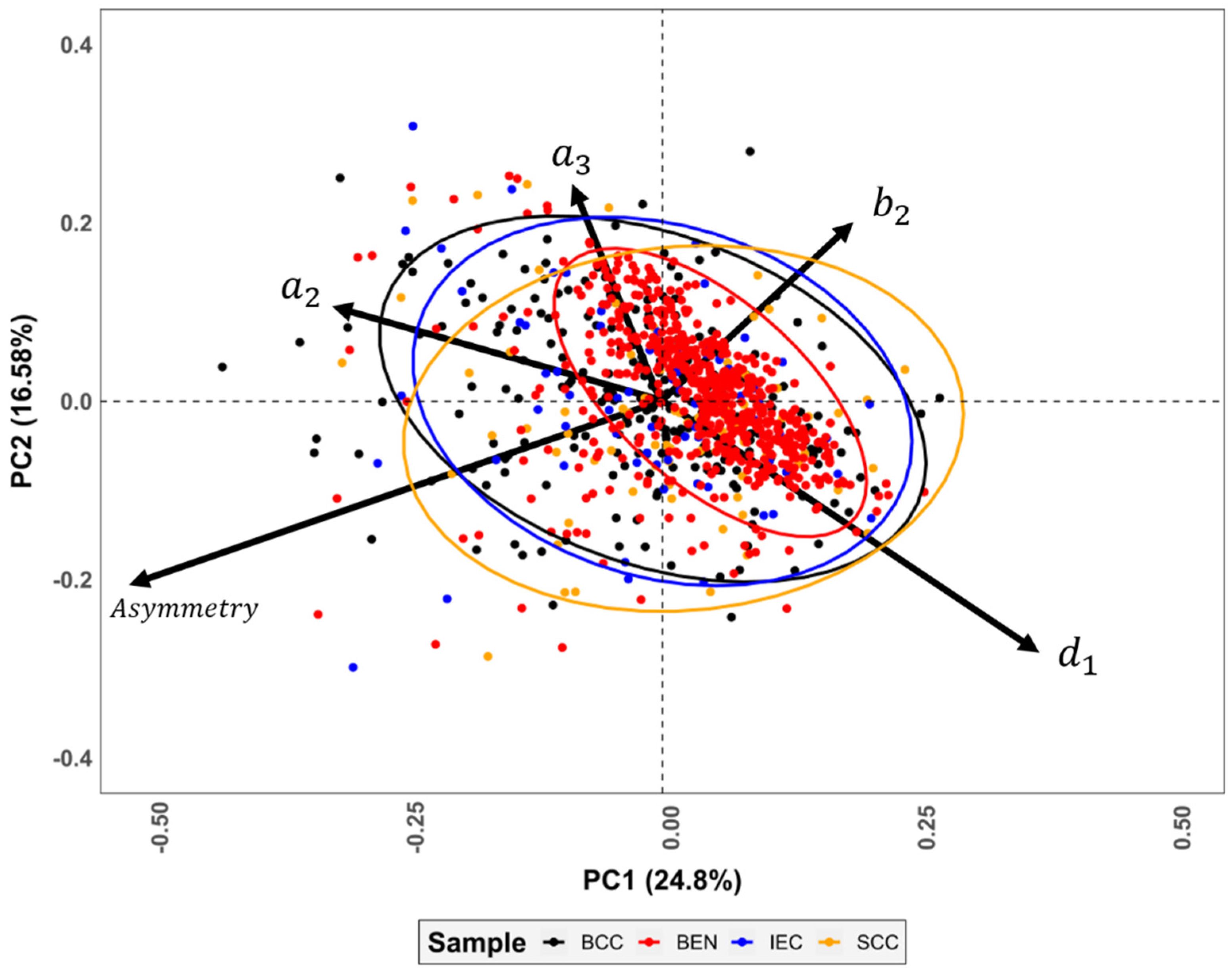
2.4. Multivariate Statistics
2.5. Asymmetry Calculations
2.6. Machine Learning
3. Experimental Results
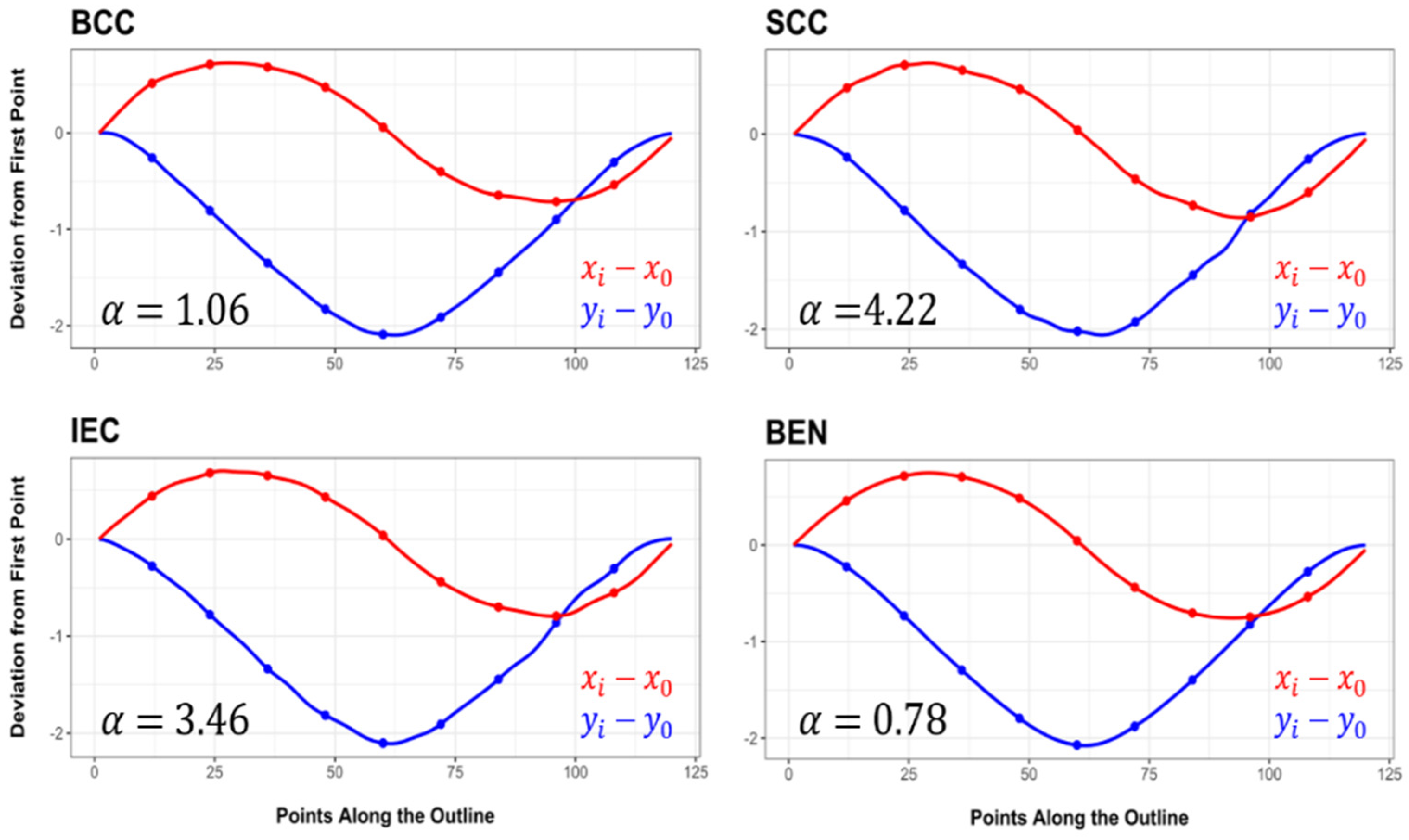
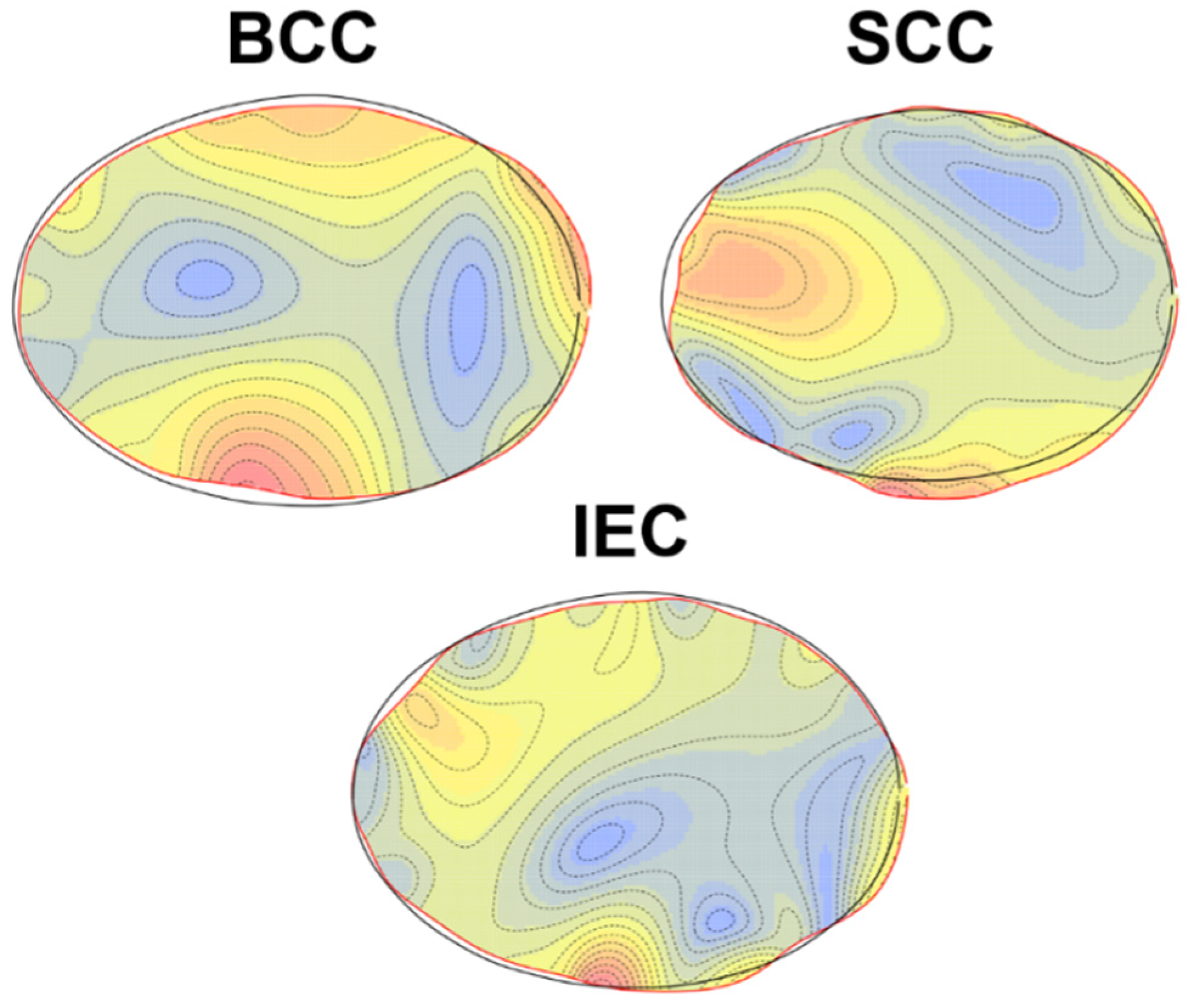
3.1. Elliptic Fourier Analyses
3.2. Lesion Asymmetry
3.3. Machine Learning
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinberg, A.S.; Ogle, C.A.; Shim, E.K. Metastatic cutaneous squamous cell carcinoma: An update. Dermatol. Surg. 2007, 33, 885–899. [Google Scholar] [CrossRef]
- Hoorens, I.; Vossaert, K.; Ongenae, K.; Brochez, L. Is early detection of basal cell carcinoma worthwhile? Systematic review based on the WHO criteria for screening. Br. J. Dermatol. 2016, 174, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.J.; Rigel, D.S.; Kopf, A.W. Early detection of malignant melanoma: The role of physician examination and self-examination of the skin. CA Cancer J. Clin. 1985, 35, 130–151. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.; Olazagasti, J.M.; Cordoro, K.M.; Brewer, J.D.; Taylor, S.C.; Bordeaux, J.S.; Chren, M.M.; Sober, A.J.; Tegeler, C.; Bhusan, R.; et al. Early detection of melanoma: Reviewing the ABCDEs. J. Am. Acad. Dermatol. 2015, 72, 717–723. [Google Scholar] [CrossRef] [PubMed]
- MacKie, R.M. An Illustrated Guide to the Recognition of Early Malignant Melanoma; University of Glasgow: Glasgow, Scotland, 1986. [Google Scholar]
- Fargnoli, M.C.; Kostaki, D.; Micantonio, T. Dermoscopy in the diagnosis and management of non-melanoma skin cancers. Artic. Eur. J. Dermatol. 2012, 22, 456–463. [Google Scholar] [CrossRef]
- Ballerini, L.; Fisher, R.B.; Aldridge, B.; Rees, J. A Color and Texture Based Hierarchical K-NN Approach to the Classification of Non-melanoma Skin Lesions. Lect. Notes Comput. Vis. Biomech. 2013, 6, 63–86. [Google Scholar]
- Courtenay, L.; Gonzalez-Aguilera, D.; Lagüela, S.; del Pozo, S.; Ruiz Méndez, C.; Barbero-García, I.; Román-Curto, C.; Cañueto, J.; Santos-Durán, C.; Cardeñoso-Álvarez, M.E.; et al. Hyperspectral Imaging and Robust Statistics in Non-Melanoma Skin Cancer Analysis. Biomed. Opt. Express 2021, 12, 5107–5127. [Google Scholar] [CrossRef] [PubMed]
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef]
- Weber, G.W.; Bookstein, F.L. Virtual Anthropology: A Guide to a New Interdisciplinary Field; Springer: Vienna, Austria, 2011. [Google Scholar]
- Bookstein, F.L. Morphometric Tools for Landmark Data; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Bookstein, F.L. Landmark Methods for Forms without Landmarks: Morphometrics of Group Differences in Outline Shape. Med. Image Anal. 1997, 1, 225–243. [Google Scholar] [CrossRef]
- Dryden, I.L.; Mardia, K.V. Statistical Shape Analysis: Wiley Series in Probability and Statistics; John Wiley Sons Ltd.: New York, NY, USA, 1998. [Google Scholar]
- Kieser, J.; Bernal, V.; Gonzalez, P.; Birch, W.; Turmaine, M.; Ichim, I. Analysis of experimental cranial skin wounding from screwdriver trauma. Int. J. Leg. Med. 2008, 122, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Komo, L.; Grassberger, M. Experimental sharp force injuries to ribs: Multimodal morphological and geometric morphometric analyses using micro-CT, macro photography and SEM. Forensic Sci. Int. 2018, 288, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Aramendi, J.; Maté-González, M.A.; Yravedra, J.; Ortega, M.C.; Arriaza, M.C.; González-Aguilera, D.; Baquedano, E.; Domínguez-Rodrigo, M. Discerning carnivore agency through the three-dimensional study of tooth pits: Revisiting crocodile feeding behaviour at FLK- Zinj and FLK NN3 (Olduvai Gorge, Tanzania). Palaeogeogr. Palaeoclim. Palaeoecol. 2017, 488, 93–102. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Huguet, R.; González-Aguilera, D.; Yravedra, J. A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification. Appl. Sci. 2020, 10, 150. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Huguet, R.; Yravedra, J. Scratches and grazes: A detailed microscopic analysis of trampling phenomena. J. Microsc. 2020, 277, 107–117. [Google Scholar] [CrossRef]
- Gunz, P.; Mitteroecker, P.; Bookstein, F.L. Semilandmarks in Three Dimensions. In Modern Morphometrics in Physical Anthropology; Slice, D.E., Ed.; Springer: Boston, MA, USA, 2005; pp. 73–98. [Google Scholar]
- Rohlf, F.J.; Archie, J.W. A Comparison of Fourier Methods for the Description of Wing Shape in Mosquitoes (Diptera: Culicidae). Syst. Biol. 1984, 33, 302–317. [Google Scholar] [CrossRef]
- Ferson, S.; Rohlf, F.J.; Koehn, R.K. Measuring Shape Variation of Two-dimensional Outlines. Syst. Biol. 1985, 34, 59–68. [Google Scholar] [CrossRef]
- Rohlf, F.J. Relationships among eigenshape analysis, Fourier analysis, and analysis of coordinates. Math. Geol. 1986, 18, 845–854. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Sinha, N.R. Evolutionary and Environmental Forces Sculpting Leaf Development. Curr. Biol. 2016, 26, 297–306. [Google Scholar] [CrossRef]
- Caple, J.; Byrd, J.; Stephan, C.N. Elliptical Fourier analysis: Fundamentals, applications, and value for forensic anthropology. Int. J. Leg. Med. 2017, 131, 1675–1690. [Google Scholar] [CrossRef]
- Ioviţă, R. Comparing Stone Tool Resharpening Trajectories with the Aid of Elliptical Fourier Analysis. In New Perspectives on Old Stones; Springer: New York, NY, USA, 2010; pp. 235–253. [Google Scholar]
- Chitwood, D.H. Imitation, Genetic Lineages, and Time Influenced the Morphological Evolution of the Violin. PLoS ONE 2014, 9, e109229. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Adhikari, A.; Roy, M. Malignant Melanoma Classification Using Cross-Platform Dataset with Deep Learning CNN Architecture. Adv. Intell. Syst. Comput. 2019, 922, 31–41. [Google Scholar]
- Izadi, S.; Mirikharaji, Z.; Kawahara, J.; Hamarneh, G. Generative adversarial networks to segment skin lesions. Proc. Int. Symp. Biomed. Imaging 2018, 15, 881–884. [Google Scholar]
- Lloyd, S.P. Least Squares Quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Serra, J. Image Analysis and Mathematical Morphology; Academic Press: New York, NY, USA, 1982. [Google Scholar]
- Jungers, W.L.; Falsetti, A.B.; Wall, C.E. Shape, relative size, and size-adjustments in morphometrics. Am. J. Phys. Anthropol. 1995, 38, 137–161. [Google Scholar] [CrossRef]
- Zahn, C.T.; Roskies, R.Z. Fourier Descriptors for Plane Closed Curves. IEEE Trans. Comput. 1972, 21, 269–281. [Google Scholar] [CrossRef]
- Giardina, C.R.; Kuhl, F.P. Accuracy of curve approximation by harmonically related vectors with elliptical loci. Comput. Graph. Image Process. 1977, 6, 277–285. [Google Scholar] [CrossRef]
- Kuhl, F.P.; Giardina, C.R. Elliptic Fourier features of a closed contour. Comput. Graph. Image Process. 1982, 18, 236–258. [Google Scholar] [CrossRef]
- Mohd Razali, N.; Bee Wah, Y. Power comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling tests. J. Stat. Model. Anal. 2011, 2, 21–33. [Google Scholar]
- Höhle, J.; Höhle, M. Accuracy assessment of digital elevation models by means of robust statistical methods. ISPRS J. Photogramm. Remote Sens. 2009, 64, 398–406. [Google Scholar] [CrossRef]
- Hotelling, H.A.; Generalized, T. Test and Measure of Multivariate Dispersion. In Proceedings of the Second Berkeley Symposium on Mathematical Statistics and Probability; Neyman, J., Ed.; University of California Press: Berkeley, CA, USA, 1951; pp. 23–42. [Google Scholar]
- Rao, C.R. An asymptotic expansion of the distribution of Wilk’s criterion. Bull. L’institut. Int. Stat. 1951, 33, 177–180. [Google Scholar]
- Hollander, M.; Wolfe, D.A. Nonparametric Statistical Methods; John Wiley & Sons: New York, NY, USA, 1973. [Google Scholar]
- Bookstein, F.L. Principal Warps: Thin-Plate Splines and the Decomposition of Deformations. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 567–585. [Google Scholar] [CrossRef]
- Wasserstein, R.L.; Lazar, N.A. The ASA Statement on p-Values: Context, Process, and Purpose. Am. Stat. 2016, 70, 129–133. [Google Scholar] [CrossRef]
- Wasserstein, R.L.; Schirm, A.L.; Lazar, N.A. Moving to a World Beyond “p < 0.05”. Am. Stat. 2019, 73, 1–19. [Google Scholar]
- Colquhoun, D. The False Positive Risk: A Proposal Concerning What to Do About p-Values. Am. Stat. 2019, 73, 192–201. [Google Scholar] [CrossRef]
- Benjamin, D.J.; Berger, J.O. Three Recommendations for Improving the Use of p-Values. Am. Stat. 2019, 73, 186–191. [Google Scholar] [CrossRef]
- Sellke, T.; Bayarri, M.J.; Berger, J.O. Calibration of p Values for Testing Precise Null Hypotheses. Am. Stat. 2012, 55, 62–71. [Google Scholar] [CrossRef]
- Colquhoun, D. The reproducibility of research and the misinterpretation of p-values. R. Soc. Open Sci. 2017, 4, 171085. [Google Scholar] [CrossRef]
- Bonhomme, V.; Picq, S.; Gaucherel, C.; Claude, J. Momocs: Outline analysis using R. J. Stat. Softw. 2014, 56, 1–24. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Herranz-Rodrigo, D.; Huguet, R.; Maté-González, M.Á.; González-Aguilera, D.; Yravedra, J. Obtaining new resolutions in carnivore tooth pit morphological analyses: A methodological update for digital taphonomy. PLoS ONE 2020, 15, e0240328. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. arXiv 2012, arXiv:1203.2944. [Google Scholar]
- Shahriari, B.; Swersky, K.; Wang, Z.; Adams, R.P.; De Freitas, N. Taking the human out of the loop: A review of Bayesian optimization. Proc. IEEE 2016, 104, 148–175. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization Yoshua Bengio. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Bergstra, J.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for Hyper-Parameter Optimization. Int. Conf. Neural. Inf. Process. Syst. 2011, 24, 2546–2554. [Google Scholar]
- He, H.; Ma, Y. Imbalanced Learning; IEEE Press: Piscataway, NJ, USA, 2013. [Google Scholar]
- Roussos, P.; Mitsea, A.; Halazonetis, D.; Sifakakis, I. Craniofacial shape in patients with beta thalassaemia: A geometric morphometric analysis. Sci. Rep. 2021, 11, 1686. [Google Scholar] [CrossRef]
- Pucciarelli, V.; Bertoli, S.; Codari, M.; de Amicis, R.; De Giorgis, V.; Battezzati, A.; Veggiotti, P.; Sforza, C. The face of Glut1-DS patients: A 3D Craniofacial Morphometric Analysis. Clin. Anat. 2017, 30, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mutsvangwa, T.E.M.; Meintjes, E.M.; Viljoen, D.L.; Douglas, T.S. Morphometric analysis and classification of the facial phenotype associated with fetal alcohol syndrome in 5- and 12-year-old children. Am. J. Med. Genet. Part. A 2010, 152, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Turam Ozdemir, S.; Ercan, I.; Ezgi Cam, F.; Ocakoglu, G.; Demirdogen, E.; Ursavas, A. Three-Dimensional Analysis of Craniofacial Shape in Obstructive Sleep Apnea Syndrome Using Geometric Morphometrics Análisis. Int. J. Morphol. 2019, 37, 338–343. [Google Scholar] [CrossRef]
- Starbuck, J.M.; Cole, T.M.; Reeves, R.H.; Richtsmeier, J.T. The Influence of trisomy 21 on facial form and variability. Am. J. Med. Genet. Part. A 2017, 173, 2861–2872. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Más, J.; Bueno-Crespo, A.; Khazendar, S.; Remezal-Solano, M.; Martínez-Cendán, J.P.; Jassim, S.; Du, H.; Al Assam, H.; Bourne, T.; Timmerman, D. Evaluation of machine learning methods with Fourier Transform features for classifying ovarian tumors based on ultrasound images. PLoS ONE 2019, 14, e0219388. [Google Scholar] [CrossRef] [PubMed]
- Sanfillipo, P.G.; Grimm, J.L.; Flanagan, J.G.; Lathrop, K.L.; Sigal, I.A. Application of Elliptic Fourier Analysis to describe the Lamina Cribrosa Shape with age and intraocular pressure. Exp. Eye Res. 2014, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leon, R.; Martinez-Vega, B.; Fabelo, H.; Ortega, S.; Melian, V.; Castaño, I.; Carretero, G.; Elmeida, P.; Garcia, A.; Quevedo, E.; et al. Non-Invasive Skin Cancer Diagnosis Using Hyperspectral Imaging for In-Situ Clinical Support. J. Clin. Med. 2020, 9, 1662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Moy, A.J.; Feng, X.; Nguyen, H.T.M.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Diffuse reflectance spectroscopy as a potential method for nonmelanoma skin cancer margin assessment. Transl. Biophotonics 2020, 2, e202000001. [Google Scholar] [CrossRef]




| MANOVA | Mahalanobis Distances | ||||||
|---|---|---|---|---|---|---|---|
| BCC | BEN | IEC | BCC | BEN | IEC | ||
| BEN | p-Value | 0.001 | 9.7 × 10−47 | ||||
| FPR | 1.8% | 2.8 × 10−42% | |||||
| IEC | p-Value | 0.756 | 0.004 | 0.228 | 1.6 × 10−27 | ||
| FPR | - | 5.7% | 37.9% | 2.7 × 10−23% | |||
| SCC | p-Value | 0.030 | 0.001 | 0.023 | 0.738 | 2.9 × 10−22 | 0.292 |
| FPR | 22.2% | 1.8% | 19.1% | - | 2.9 × 10−10% | 49.4% | |
| Shapiro | ||||||||
|---|---|---|---|---|---|---|---|---|
| w | p | Min. | L (0.05) | Median | √BWMV | U (0.95) | Max. | |
| BCC * | 0.782 | 2.2 × 10−16 | 0.131 | 0.179 | 0.292 | 0.099 | 0.554 | 1.064 |
| IEC * | 0.647 | 2.6 × 10−12 | 0.168 | 0.186 | 0.292 | 0.081 | 0.621 | 1.285 |
| SCC * | 0.566 | 1.2 × 10−14 | 0.097 | 0.172 | 0.270 | 0.084 | 0.572 | 1.390 |
| BEN | 0.616 | 2.2 × 10−16 | 0.134 | 0.183 | 0.239 | 0.046 | 0.373 | 1.213 |
| Cancer * | 0.679 | 2.2 × 10−16 | 0.097 | 0.183 | 0.289 | 0.093 | 0.554 | 1.390 |
| Benign | 0.616 | 2.2 × 10−16 | 0.134 | 0.183 | 0.239 | 0.046 | 0.373 | 1.213 |
| MANOVA | Mahalanobis Distances | ||||||
|---|---|---|---|---|---|---|---|
| BCC | BEN | IEC | BCC | BEN | IEC | ||
| BEN | p-Value | 0.001 | 3.6× 10−45 | ||||
| FPR | 1.8% | 1.0 × 10−40% | |||||
| IEC | p-Value | 0.814 | 0.001 | 0.242 | 3.4 × 10−27 | ||
| FPR | - | 1.8% | 48.3% | 5.6 × 10−23% | |||
| SCC | p-Value | 0.058 | 0.001 | 0.021 | 0.452 | 4.0 × 10−23 | 0.051 |
| FPR | 31.0% | 1.8% | 18.1% | - | 5.6 × 10−19% | 29.2% | |
| Training Variables | Accuracy | Precision | Recall | F-Statistic | AUC |
|---|---|---|---|---|---|
| Asymmetry | 0.690 | 0.646 | 0.447 | 0.528 | 0.696 |
| EFA Coefficients | 0.772 | 0.887 | 0.717 | 0.794 | 0.693 |
| EFA & Asymmetry | 0.765 | 0.883 | 0.711 | 0.788 | 0.685 |
| Combined EFA & Asymmetry | 0.786 | 0.915 | 0.717 | 0.804 | 0.735 |
| True | |||
|---|---|---|---|
| Benign | Malignant | ||
| Predicted | Benign | 71.67% | 10.53% |
| Malignant | 28.33% | 89.47% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Courtenay, L.A.; Barbero-García, I.; Aramendi, J.; González-Aguilera, D.; Rodríguez-Martín, M.; Rodríguez-Gonzalvez, P.; Cañueto, J.; Román-Curto, C. A Novel Approach for the Shape Characterisation of Non-Melanoma Skin Lesions Using Elliptic Fourier Analyses and Clinical Images. J. Clin. Med. 2022, 11, 4392. https://doi.org/10.3390/jcm11154392
Courtenay LA, Barbero-García I, Aramendi J, González-Aguilera D, Rodríguez-Martín M, Rodríguez-Gonzalvez P, Cañueto J, Román-Curto C. A Novel Approach for the Shape Characterisation of Non-Melanoma Skin Lesions Using Elliptic Fourier Analyses and Clinical Images. Journal of Clinical Medicine. 2022; 11(15):4392. https://doi.org/10.3390/jcm11154392
Chicago/Turabian StyleCourtenay, Lloyd A., Innes Barbero-García, Julia Aramendi, Diego González-Aguilera, Manuel Rodríguez-Martín, Pablo Rodríguez-Gonzalvez, Javier Cañueto, and Concepción Román-Curto. 2022. "A Novel Approach for the Shape Characterisation of Non-Melanoma Skin Lesions Using Elliptic Fourier Analyses and Clinical Images" Journal of Clinical Medicine 11, no. 15: 4392. https://doi.org/10.3390/jcm11154392
APA StyleCourtenay, L. A., Barbero-García, I., Aramendi, J., González-Aguilera, D., Rodríguez-Martín, M., Rodríguez-Gonzalvez, P., Cañueto, J., & Román-Curto, C. (2022). A Novel Approach for the Shape Characterisation of Non-Melanoma Skin Lesions Using Elliptic Fourier Analyses and Clinical Images. Journal of Clinical Medicine, 11(15), 4392. https://doi.org/10.3390/jcm11154392










