Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
2.2. Image Preprocessing
2.3. Estimation of Texture Parameters
- Histogram (9 features): histogram mean, histogram variance, histogram skewness, histogram kurtosis, percentiles 1%, 10%, 50%, 90%, and 99%;
- Gradient (5 features): absolute gradient mean, absolute gradient variance, absolute gradient skewness, absolute gradient kurtosis, percentage of pixels with a nonzero gradient;
- Run length matrix (5 features × 4 various directions): run length nonuniformity, grey level nonuniformity, long run emphasis, short run emphasis, the fraction of image in runs;
- Co-occurrence matrix (11 features × 4 various directions × 5 between-pixels distances) angular second moment, contrast, correlation, sum of squares, inverse difference moment, sum average, sum variance, sum entropy, entropy, difference variance, difference entropy;
- Autoregressive model (5 features): parameters Θ1, Θ2, Θ3, Θ4, standard deviation.
- Haar wavelet (24 features): wavelet energy (features are computed at 6 scales within 4 frequency bands LL, LH, HL, and HH).
2.4. Data Preprocessing
- Removal of features with constant values (variance equal to 0).
- Removal of features with nearly constant values (variance lower than 0.01).
- Removal of duplicated features.
- Removal of correlated features
2.5. Data Reduction
- Filter methods:
- Univariate—Fisher’s method (FISHER) and variance analysis method (ANOVA);
- Multivariate—Relief method (RELIEF).
- Wrapper methods:
- Sequential Forward Selection (SFS);
- Sequential Backward Selection (SBS);
- Recursive Feature Elimination along with LogisticRegression estimator (RFE).
- Embedded methods:
- SelectFromModel meta-transformer and logistic regression estimator (LR);
- SelectFromModel meta-transformer and AdaBoost estimator (ADA);
- SelectFromModel meta-transformer along with LightGBM estimator (LGBM).
2.6. Training the Classification Models
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prakash, P.; Kalra, M.K.; Digumarthy, S.R.; Hsieh, J.; Pien, H.; Singh, S.; Gilman, M.D.; Shepard, J.-A.O. Radiation Dose Reduction with Chest Computed Tomography Using Adaptive Statistical Iterative Reconstruction Technique: Initial Experience. J. Comput. Assist. Tomogr. 2010, 34, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Lawder, H.J.; Hara, A.; Kujak, J.; Pavlicek, W. Innovations in CT Dose Reduction Strategy: Application of the Adaptive Statistical Iterative Reconstruction Algorithm. AJR Am. J. Roentgenol. 2010, 194, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.; Samei, E. Quantum Noise Properties of CT Images with Anatomical Textured Backgrounds across Reconstruction Algorithms: FBP and SAFIRE. Med. Phys. 2014, 41, 091908. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.H. Algebraic Reconstruction in CT from Limited Views. IEEE Trans. Med. Imaging 1989, 8, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Huda, W.; Ogden, K.M.; Samei, E.; Scalzetti, E.M.; Lavallee, R.L.; Roskopf, M.L.; Groat, G.E. Reconstruction Filters and Contrast Detail Curves in CT. In Proceedings of the Medical Imaging 2008: Image Perception, Observer Performance, and Technology Assessment; International Society for Optics and Photonics, San Diego, CA, USA, 6 March 2008; Volume 6917, p. 691710. [Google Scholar]
- Shahabaz; Somwanshi, D.K.; Yadav, A.K.; Roy, R. Medical Images Texture Analysis: A Review. In Proceedings of the 2017 International Conference on Computer, Communications and Electronics (Comptelix), Jaipur, India, 1–2 July 2017. [Google Scholar] [CrossRef]
- Blair, J.P.; Bierma-Zeinstra, S.; Nielsen, H.B.; Bay-Jensen, A.-C.; Dam, E.B. Texture Analysis of Knee Pathology for the Diagnosis of Radiographic Osteoarthritis. Osteoarthr. Cartil. 2018, 26, S436. [Google Scholar] [CrossRef]
- Fan, J.; Min, B.; Xu, F.; Yang, P.; Ye, H.; Feng, Y.; Shen, Y.; Xu, C.; Zhou, W.; Ren, X. Clinical Diagnosis of Pancreatic Cancer Using Texture Analysis in Endoscopic Ultrasonography. J. Med. Imaging Health Inform. 2019, 9, 1844–1848. [Google Scholar] [CrossRef]
- Fujima, N.; Homma, A.; Harada, T.; Shimizu, Y.; Tha, K.K.; Kano, S.; Mizumachi, T.; Li, R.; Kudo, K.; Shirato, H. The Utility of MRI Histogram and Texture Analysis for the Prediction of Histological Diagnosis in Head and Neck Malignancies. Cancer Imaging 2019, 19, 5. [Google Scholar] [CrossRef]
- Gilanie, G.; Bajwa, U.I.; Waraich, M.M.; Habib, Z. Computer Aided Diagnosis of Brain Abnormalities Using Texture Analysis of MRI Images. Int. J. Imaging Syst. Technol. 2019, 29, 260–271. [Google Scholar] [CrossRef]
- Li, Z.; Yu, L.; Wang, X.; Yu, H.; Gao, Y.; Ren, Y.; Wang, G.; Zhou, X. Diagnostic Performance of Mammographic Texture Analysis in the Differential Diagnosis of Benign and Malignant Breast Tumors. Clin. Breast Cancer 2018, 18, e621–e627. [Google Scholar] [CrossRef]
- Manabe, O.; Ohira, H.; Hirata, K.; Hayashi, S.; Naya, M.; Tsujino, I.; Aikawa, T.; Koyanagawa, K.; Oyama-Manabe, N.; Tomiyama, Y.; et al. Use of 18F-FDG PET/CT Texture Analysis to Diagnose Cardiac Sarcoidosis. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 1240–1247. [Google Scholar] [CrossRef]
- Mitrea, D.; Nedevschi, S.; Mitrea, P.; Brehar, R.; Vancea, F.I.; Platon(lupşor), M.; Ştefănescu, H.; Badea, R. Advanced Texture Analysis and Classification Methods for the Automatic Diagnosis of the Hepatocellular Carcinoma. Appl. Med. Inform. 2019, 41, 17–18. [Google Scholar]
- WANG Minhong, Z.L.; WANG Minhong, Z.L. Value of Conventional MRI Texture Analysis in Differential Diagnosis of Glioblastomas and Primary Central Nervous System Lymphoma. China Oncol. 2019, 29, 284–288. [Google Scholar] [CrossRef]
- Wang, X.; Suo, S.; Zhu, J.; Feng, Q.; Hua, X.; Wang, W.; Gong, J.; Liu, J.; Lu, Q. Application of intestinal CT texture analysis and nonlinear discriminant analysis in differential diagnosis of colorectal cancer and ulcerative colitis. J. Shanghai Jiaotong Univ. 2018, 38, 624. [Google Scholar] [CrossRef]
- Masci, G.M.; Ciccarelli, F.; Mattei, F.I.; Grasso, D.; Accarpio, F.; Catalano, C.; Laghi, A.; Sammartino, P.; Iafrate, F. Role of CT Texture Analysis for Predicting Peritoneal Metastases in Patients with Gastric Cancer. Radiol. Med. 2022, 127, 251–258. [Google Scholar] [CrossRef]
- Nagawa, K.; Suzuki, M.; Yamamoto, Y.; Inoue, K.; Kozawa, E.; Mimura, T.; Nakamura, K.; Nagata, M.; Niitsu, M. Texture Analysis of Muscle MRI: Machine Learning-Based Classifications in Idiopathic Inflammatory Myopathies. Sci. Rep. 2021, 11, 9821. [Google Scholar] [CrossRef]
- Cavallo, A.U.; Troisi, J.; Forcina, M.; Mari, P.-V.; Forte, V.; Sperandio, M.; Pagano, S.; Cavallo, P.; Floris, R.; Garaci, F. Texture Analysis in the Evaluation of COVID-19 Pneumonia in Chest X-Ray Images: A Proof of Concept Study. Curr. Med. Imaging Rev. 2021, 17, 1094–1102. [Google Scholar] [CrossRef]
- Downey, P.A.; Siegel, M.I. Bone Biology and the Clinical Implications for Osteoporosis. Phys. Ther. 2006, 86, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Marcus, R.; Dempster, D.; Cauley, J.; Feldman, D. Osteoporosis, 4th ed.; Academic Press: Cambridge, MA, USA, 2013; ISBN 978-0-12-415853-5. [Google Scholar]
- Ebeling, P.R.; Nguyen, H.H.; Aleksova, J.; Vincent, A.J.; Wong, P.; Milat, F. Secondary Osteoporosis. Endocr. Rev. 2022, 43, 240–313. [Google Scholar] [CrossRef]
- Kanis, J.A. Diagnosis of Osteoporosis and Assessment of Fracture Risk. Lancet 2002, 359, 1929–1936. [Google Scholar] [CrossRef]
- Klibanski, A.; Adams-Campbell, L.; Bassford, T.; Blair, S.N.; Boden, S.D.; Dickersin, K.; Gifford, D.R.; Glasse, L.; Goldring, S.R.; Hruska, K.; et al. Osteoporosis Prevention, Diagnosis, and Therapy. JAMA 2001, 285, 785–795. [Google Scholar]
- Orimo, H.; Hayashi, Y.; Fukunaga, M.; Sone, T.; Fujiwara, S.; Shiraki, M.; Kushida, K.; Miyamoto, S.; Soen, S.; Nishimura, J.; et al. Diagnostic Criteria for Primary Osteoporosis: Year 2000 Revision. J. Bone Miner. Metab. 2001, 19, 331–337. [Google Scholar] [CrossRef]
- Mustapha, A.; Hussain, A.; Zulkifley, M.A.; Zaki, W.M.D.W.; Husain, H.; Abdul Hamid, H.B. Anterior Osteoporosis Classification in Cervical Vertebrae Using Fuzzy Decision Tree. Multimed. Tools Appl. 2017, 77, 4011–4045. [Google Scholar] [CrossRef]
- Mustapha, A.; Hussain, A.; Samad, S.; Zulkifley, M.; Diyana Wan Zaki, W.; Hamid, H. Design and Development of a Content-Based Medical Image Retrieval System for Spine Vertebrae Irregularity. Biomed. Eng. Online 2015, 14, 6. [Google Scholar] [CrossRef] [Green Version]
- Stanley, R.J.; Antani, S.; Long, R.; Thoma, G.; Gupta, K.; Das, M. Size-Invariant Descriptors for Detecting Regions of Abnormal Growth in Cervical Vertebrae. Comput. Med. Imaging Graph. 2008, 32, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Lespessailles, E.; Gadois, C.; Kousignian, I.; Neveu, J.P.; Fardellone, P.; Kolta, S.; Roux, C.; Do-Huu, J.P.; Benhamou, C.L. Clinical Interest of Bone Texture Analysis in Osteoporosis: A Case Control Multicenter Study. Osteoporos. Int. 2008, 19, 1019–1028. [Google Scholar] [CrossRef]
- Gaidel, A.; Khramov, A. Application of Texture Analysis for Automated Osteoporosis Diagnostics by Plain Hip Radiography. Pattern Recognit. Image Anal. 2015, 25, 301–305. [Google Scholar] [CrossRef]
- Chappard, D.; Pascaretti-Grizon, F.; Gallois, Y.; Mercier, P.; Baslé, M.F.; Audran, M. Medullar Fat Influences Texture Analysis of Trabecular Microarchitecture on X-Ray Radiographs. Eur. J. Radiol. 2006, 58, 404–410. [Google Scholar] [CrossRef]
- Lespessailles, E.; Gadois, C.; Lemineur, G.; Do-Huu, J.P.; Benhamou, L. Bone Texture Analysis on Direct Digital Radiographic Images: Precision Study and Relationship with Bone Mineral Density at the Os Calcis. Calcif. Tissue Int. 2007, 80, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Bar, A.; Wolf, L.; Bergman Amitai, O.; Toledano, E.; Elnekave, E. Compression Fractures Detection on CT. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis, Orlando, Fl, USA, 3 March 2017; Armato, S.G., Petrick, N.A., Eds.; SPIE: Bellingham, WC, USA, 2017; Volume 10134. [Google Scholar]
- Roth, H.R.; Wang, Y.; Yao, J.; Lu, L.; Burns, J.E.; Summers, R.M. Deep Convolutional Networks for Automated Detection of Posterior-Element Fractures on Spine CT. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, International Society for Optics and Photonics, San Diego, CA, USA, 24 March 2016; SPIE: Bellingham, WC, USA, 2016; Volume 9785, p. 97850. [Google Scholar]
- Emohare, O.; Cagan, A.; Morgan, R.; Davis, R.; Asis, M.; Switzer, J.; Polly, D.W. The Use of Computed Tomography Attenuation to Evaluate Osteoporosis Following Acute Fractures of the Thoracic and Lumbar Vertebra. Geriatr. Orthop. Surg. Rehabil. 2014, 5, 50–55. [Google Scholar] [CrossRef]
- Dzierżak, R.; Omiotek, Z.; Tkacz, E.; Kępa, A. The Influence of the Normalisation of Spinal CT Images on the Significance of Textural Features in the Identification of Defects in the Spongy Tissue Structure. In Proceedings of the Innovations in Biomedical Engineering, Katowice, Poland, 18–20 October 2018; Tkacz, E., Gzik, M., Paszenda, Z., Piętka, E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 55–66. [Google Scholar]
- Oprogramowanie --> Program MaZda. Available online: http://www.eletel.p.lodz.pl/programy/cost/progr_mazda.html (accessed on 3 May 2020).
- Haralick, R.M. Statistical and Structural Approaches to Texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Dennis, T.J. Textured Image Segmentation by Context Enhanced Clustering. IEE Proc. Vis. Image Signal Process. 1994, 141, 413–421. [Google Scholar] [CrossRef]
- Lerski, R.A.; Straughan, K.; Schad, L.R.; Boyce, D.; Blüml, S.; Zuna, I. MR Image Texture Analysis—An Approach to Tissue Characterization. Magn. Reson. Imaging 1993, 11, 873–887. [Google Scholar] [CrossRef]
- Omiotek, Z. Improvement of the Classification Quality in Detection of Hashimoto’s Disease with a Combined Classifier Approach. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2017, 231, 774–782. [Google Scholar] [CrossRef]
- Omiotek, Z.; Stepanchenko, O.; Wójcik, W.; Legieć, W.; Szatkowska, M. The Use of the Hellwig’s Method for Feature Selection in the Detection of Myeloma Bone Destruction Based on Radiographic Images. Biocybern. Biomed. Eng. 2019, 39, 328–338. [Google Scholar] [CrossRef]
- Dzierżak, R. Comparison of the influence of standardization and normalization of data on the effectiveness of spongy tissue texture classification. Inform. Control Meas. Econ. Environ. Prot. 2019, 9, 66–69. [Google Scholar] [CrossRef]
- Scikit-Learn: Machine Learning in Python—Scikit-Learn 0.22.2 Documentation. Available online: https://scikit-learn.org/stable/ (accessed on 3 May 2020).
- Hassani, A.S.E.B.E.; Hassouni, M.E.; Houam, L.; Rziza, M.; Lespessailles, E.; Jennane, R. Texture Analysis Using Dual Tree M-Band and Rényi Entropy. Application to Osteoporosis Diagnosis on Bone Radiographs. In Proceedings of the 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 2–5 May 2012. [Google Scholar] [CrossRef]
- Singh, A.; Dutta, M.K.; Jennane, R.; Lespessailles, E. Classification of the Trabecular Bone Structure of Osteoporotic Patients Using Machine Vision. Comput. Biol. Med. 2017, 91, 148–158. [Google Scholar] [CrossRef]
- Zheng, K.; Makrogiannis, S. Bone Texture Characterization for Osteoporosis Diagnosis Using Digital Radiography. Conf Proc IEEE Eng. Med. Biol. Soc. 2016, 2016, 1034–1037. [Google Scholar] [CrossRef] [Green Version]
- Areeckal, A.S.; Kamath, J.; Zawadynski, S.; Kocher, M.; Sumam, D.S. Combined Radiogrammetry and Texture Analysis for Early Diagnosis of Osteoporosis Using Indian and Swiss Data. Comput. Med. Imaging Graph. 2018, 68, 25–39. [Google Scholar] [CrossRef]
- Omiotek, Z.; Dzierżak, R.; Uhlig, S. Fractal Analysis of the Computed Tomography Images of Vertebrae on the Thoraco-Lumbar Region in Diagnosing Osteoporotic Bone Damage. Proc. Inst. Mech. Eng. H 2019, 233, 1269–1281. [Google Scholar] [CrossRef]
- Yger, F. Challenge IEEE-ISBI/TCB: Application of Covariance Matrices and Wavelet Marginals. arXiv 2014, arXiv:1410.2663. [Google Scholar]
- Houam, L.; Hafiane, A.; Boukrouche, A.; Lespessailles, E.; Jennane, R. One Dimensional Local Binary Pattern for Bone Texture Characterization. Pattern Anal. Appl. 2014, 17, 179–193. [Google Scholar] [CrossRef]
- Tafraouti, A.; Hassouni, M.E.; Toumi, H.; Lespessailles, E.; Jennane, R. Osteoporosis Diagnosis Using Frequency Separation and Fractional Brownian Motion. In Proceedings of the 2017 International Conference on Wireless Networks and Mobile Communications (WINCOM), Rabat, Morocco, 1–4 November 2017; pp. 1–4. [Google Scholar]
- Oulhaj, H.; Rziza, M.; Amine, A.; Toumi, H.; Lespessailles, E.; El Hassouni, M.; Jennane, R. Anisotropic Discrete Dual-Tree Wavelet Transform for Improved Classification of Trabecular Bone. IEEE Trans. Med. Imaging 2017, 36, 2077–2086. [Google Scholar] [CrossRef]
- Oulhaj, H.; Rziza, M.; Amine, A.; Toumi, H.; Lespessailles, E.; Jennane, R.; Hassouni, M.E. Trabecular Bone Characterization Using Circular Parametric Models. Biomed. Signal Process. Control 2017, 33, 411–421. [Google Scholar] [CrossRef]
- Harrar, K.; Jennane, R.; Zaouchi, K.; Janvier, T.; Toumi, H.; Lespessailles, E. Oriented Fractal Analysis for Improved Bone Microarchitecture Characterization. Biomed. Signal Process. Control 2018, 39, 474–485. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.W.; Jeong, J.-W.; Yoo, D.-S.; Kim, S. A Preliminary Study on Discrimination of Osteoporotic Fractured Group from Nonfractured Group Using Support Vector Machine. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008, 2008, 474–477. [Google Scholar] [CrossRef]
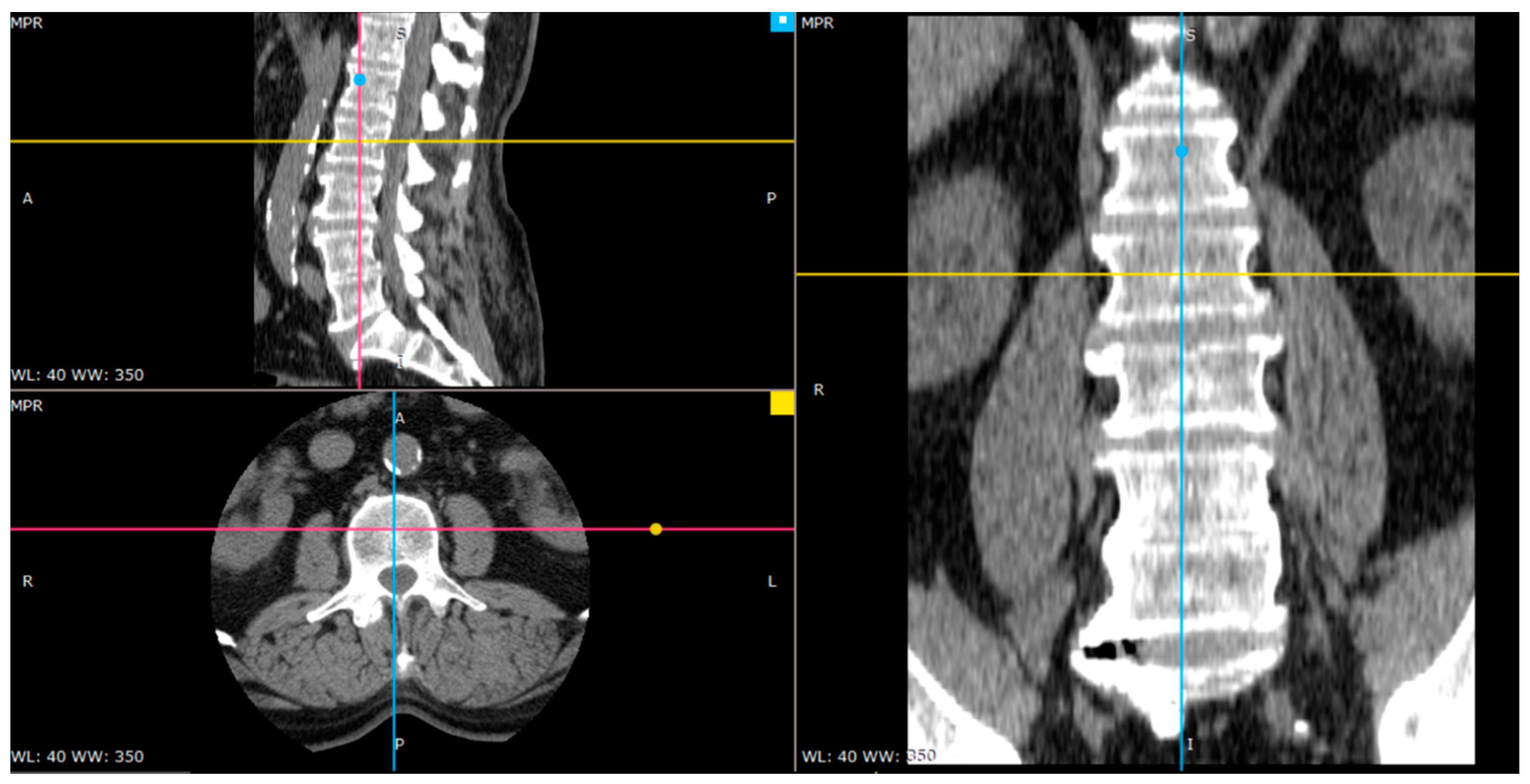
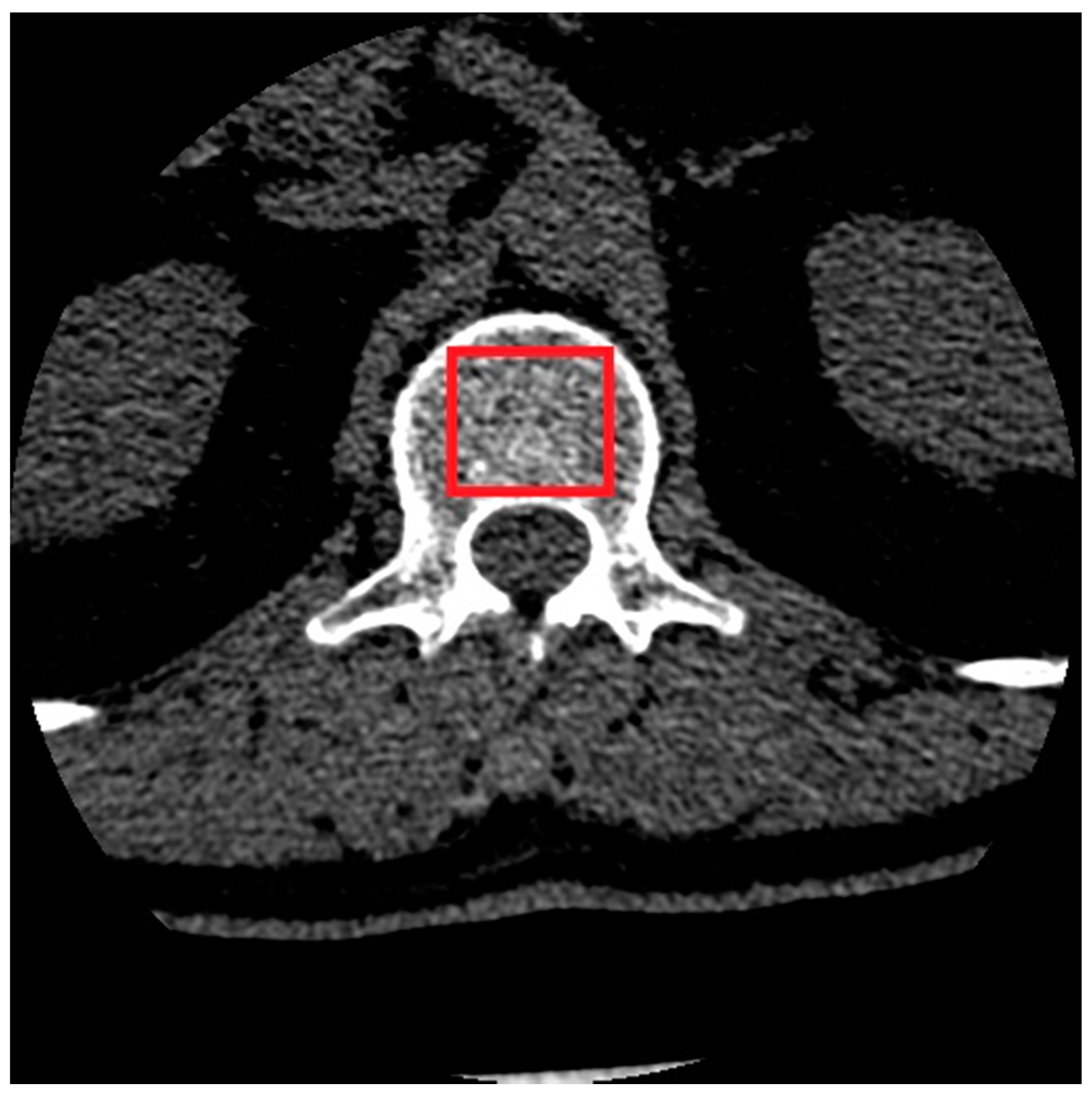
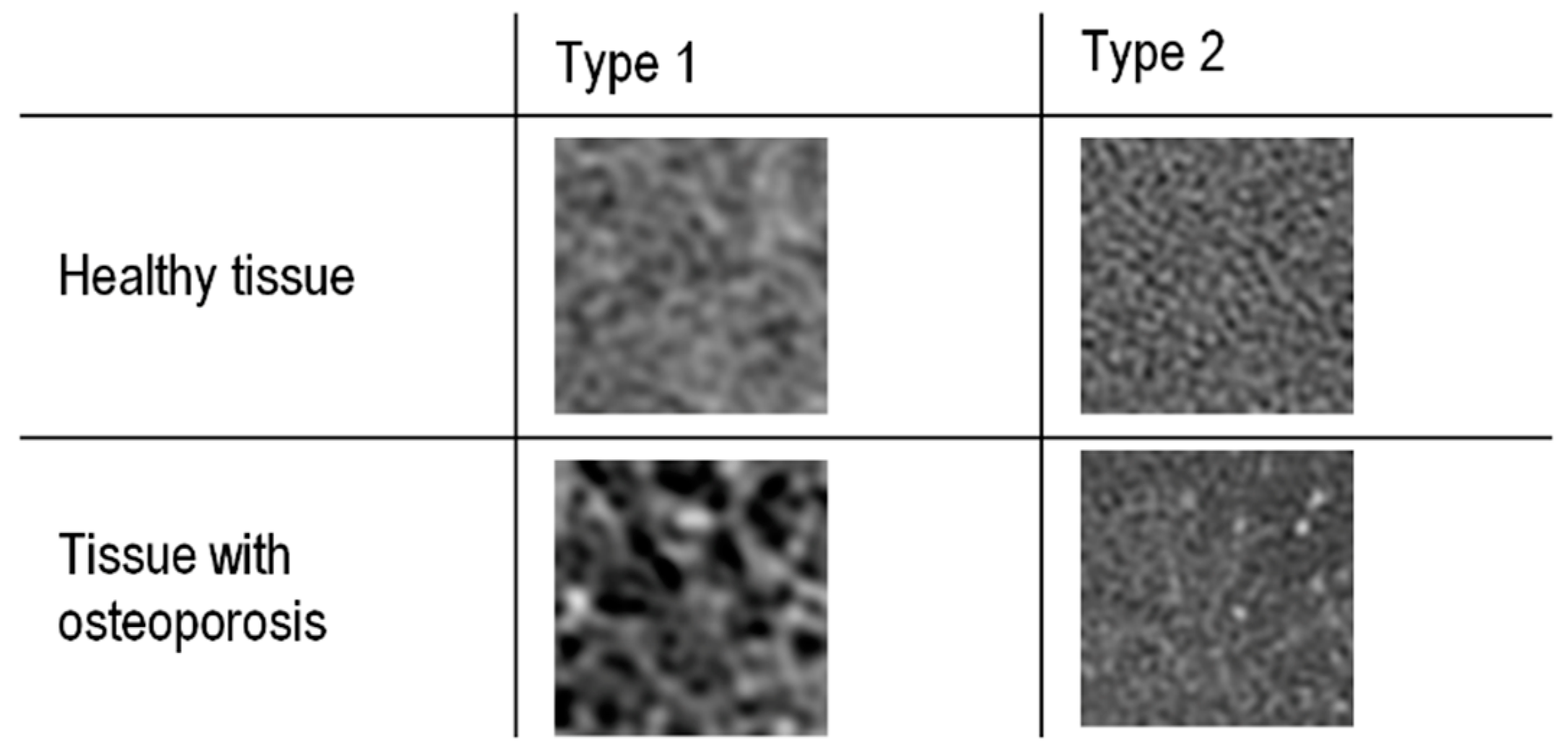
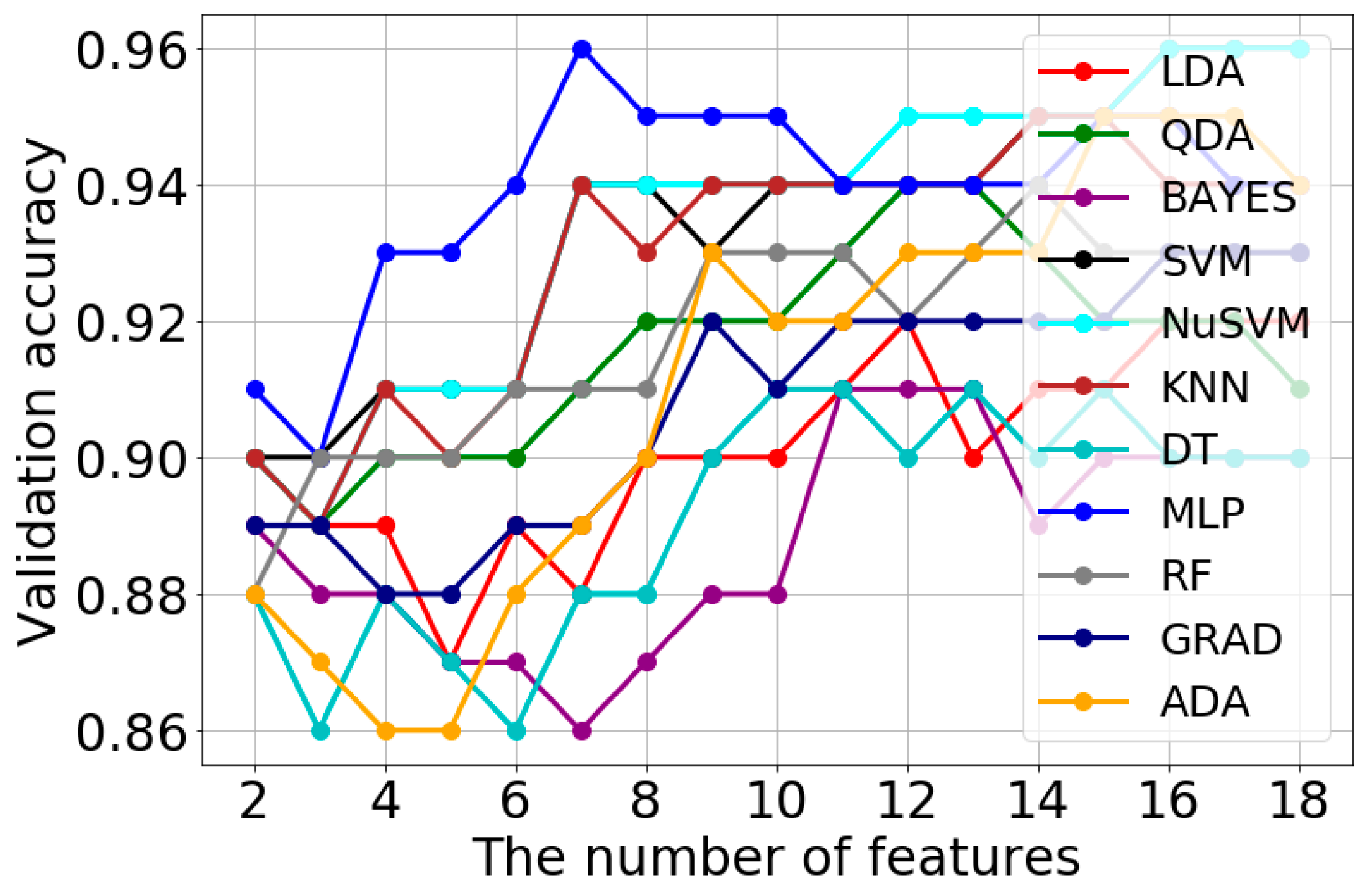

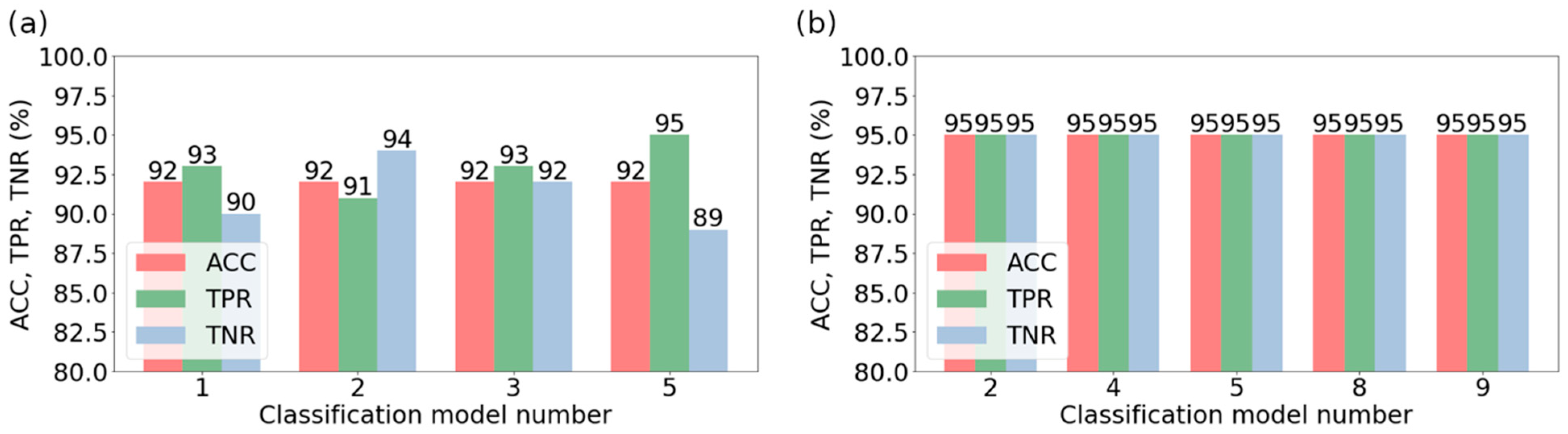
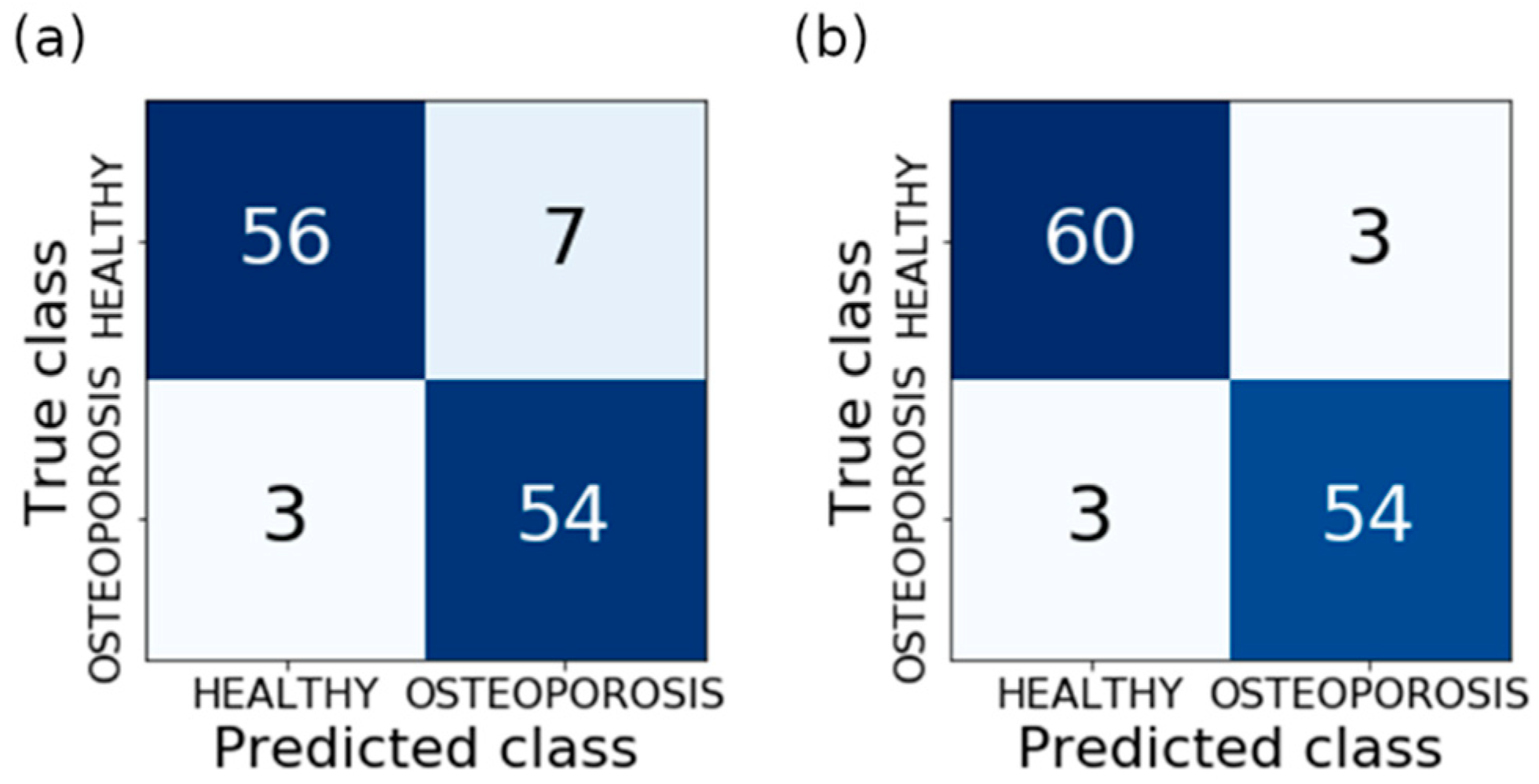
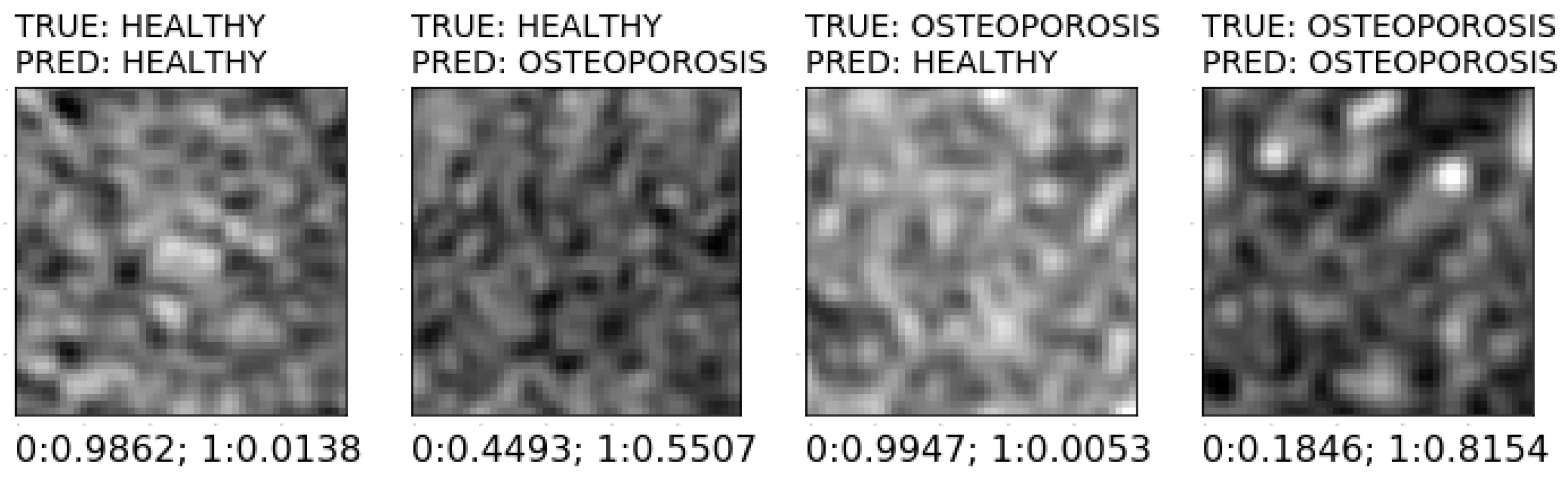
| Classification Method | Validation Accuracy | Optimal Features Number | Optimal Model Parameters |
|---|---|---|---|
| LDA | 0.92 | 12 | solver = ‘svd’ |
| QDA | 0.94 | 12 | tol = 1 × 10−5 |
| BAYES | 0.91 | 11 | var_smoothing = 0.1 |
| SVM | 0.96 | 16 | C = 1.0, gamma = ‘scale’, kernel = ‘rbf’ |
| NuSVM | 0.96 | 16 | gamma = ‘scale’, kernel = ‘rbf’, nu = 0.3 |
| KNN | 0.95 | 14 | n_neighbors = 5 |
| DT | 0.91 | 10 | criterion = ‘gini’, max_depth = 3 |
| MLP | 0.96 | 7 | activation = ‘relu’, alpha = 0.1, solver = ‘lbfgs’, max_iter = 1000, hidden_layer_sizes = (3,) |
| RF | 0.94 | 14 | max_depth = 5, n_estimators = 20 |
| GRAD | 0.93 | 16 | loss = ‘deviance’, n_estimators = 50 |
| ADA | 0.95 | 15 | n_estimators = 50 |
| Model Number | Bone Reconstruction | Soft Tissue Reconstruction | ||||||
|---|---|---|---|---|---|---|---|---|
| Classification Method | Feature Selection Method | The Number of Features | Validation Accuracy (%) | Classification Method | Feature Selection Method | The Number of Features | Validation Accuracy (%) | |
| 1 | NuSVM | FISHER | 13 | 94 | MLP | FISHER | 7 | 96 |
| 2 | RF | ANOVA | 26 | 94 | NuSVM | ANOVA | 17 | 96 |
| 3 | SVM | RELIEF | 27 | 94 | MLP | ANOVA | 17 | 96 |
| 4 | NuSVM | SFS | 6 | 94 | SVM | RELIEF | 18 | 96 |
| 5 | KNN | SBS | 9 | 94 | NuSVM | RELIEF | 18 | 96 |
| 6 | RF | SBS | 9 | 94 | KNN | SFS | 5 | 96 |
| 7 | MLP | RFE | 18 | 95 | KNN | SBS | 5 | 96 |
| 8 | ADA | 9 | 93 | SVM | RFE | 18 | 96 | |
| 9 | LR | 10 | 93 | NuSVM | RFE | 18 | 96 | |
| 10 | LGBM | 3 | 89 | ADA | 8 | 95 | ||
| 11 | LR | 4 | 91 | |||||
| 12 | LGBM | 2 | 88 | |||||
| Model Number | Classification Method | Feature Selection Method | The Number of Features | Model Parameters |
|---|---|---|---|---|
| 2 | NuSVM | ANOVA | 17 | gamma = ‘scale’, kernel = ‘rbf’, nu = 0.3 |
| 4 | SVM | RELIEF | 18 | C = 1.0, gamma = ‘scale’, kernel = ‘rbf’ |
| 5 | NuSVM | RELIEF | 18 | gamma = ‘scale’, kernel = ‘rbf’, nu = 0.3 |
| 8 | SVM | RFE | 18 | C = 1.0, gamma = ‘scale’, kernel = ‘rbf’ |
| 9 | NuSVM | RFE | 18 | gamma = ‘scale’, kernel = ‘rbf’, nu = 0.3 |
| No. in Ref. | Texture Features | ROI Segmentation | Dataset | Classifier | TPR | TNR | PPV | NPV | ACC | F1-Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Own results | Histogram, Gradient, Run length matrix, Cooccurrence, Autoregressive, Haar wavelet | Manual | 50 cases & 50 controls | SVM NuSVM | 95 | 95 | - | - | 95 | - |
| [49] | power spectral density, triangular prism surface area and variation, box counting, | Manual | 11 cases & 13 controls | K-NN | 78 | 90 | 90 | 77 | 81 | - |
| [50] | Wavelet Marginals-Haar | Calcaneal (Manual) | 58 cases & 58 controls | SVM | 62.1 | 65.5 | 64.3 | 63.3 | 63.8 | 63.2 |
| [51] | 1D LBP | Calcaneal (Manual) | 39 cases & 41 controls | KNN | - | 43.9 | - | - | 71.3 | 77.2 |
| [47] | Fractal dimension, wavelet analysis, Gabor, LBP, DFT, DCT, Laws masks, edge histogram and GLCM | Calcaneal (Manual) | 58 cases & 58 controls | RF | 74.1 | 74.1 | - | - | 74.1 | - |
| [52] | 1D projection modeled as fractional Brownian motion | Calcaneal (Manual) | - | SVM | 96.9 | 97.6 | - | - | 94.5 | 94.3 |
| [45] | Fractional Brownian model and Rao geodesic distance | Calcaneal | 348 cases & 348 controls | KNN | 97.8 | 95.4 | - | - | 96.6 | 96.5 |
| [46] | Histogram and GLCM and PCA analysis | Calcaneal (Manual) | 87 cases & 87 controls | SVM | 97.7 | 95.4 | 95.5 | 97.7 | 96.6 | 96.6 |
| [53] | Anisotropic discrete dual-tree wavelet transform | Calcaneal (Manual) | 87 cases & 87 controls | SVM | - | 93.1 | 92.9 | 91.0 | 91.9 | 91.9 |
| [54] | Wavelet decomposition and parametric circular models | Calcaneal (Manual) | 87 cases & 87 controls | SVM | 100 | 92.5 | 91.9 | 100 | 95.9 | 95.8 |
| [55] | Oriental fractal analysis | Calcaneal (Manual) | 87 cases & 87 controls | - | 72.0 | 71.0 | 72.0 | 71.0 | 71.8 | 72.2 |
| [56] | BMD, fractal, histomorphometric and skeletal measures | Distal radius | 87 cases & 87 controls | SVM | 79.0 | 66.0 | - | - | - | - |
| [48] | Cortical, histogram, GLCM and MGM | Distal radius (Automated) | 60 cases & 60 controls | SVM | 86.7 | 65.0 | 71.2 | 83.0 | 75.8 | 78.2 |
| [48] | Cortical and LLBP | Distal radius (Automated) | 60 cases & 60 controls | SVM | 88.3 | 66.7 | 72.6 | 85.1 | 77.5 | 79.7 |
| [48] | Cortical and hLLBP | Distal radius (Automated) | 60 cases & 60 controls | LR | 81.7 | 76.7 | 77.8 | 80.7 | 79.2 | 79.7 |
| [48] | Cortical and vLLBP | Distal radius (Automated) | 60 cases & 60 controls | SVM | 88.3 | 60.0 | 68.8 | 83.7 | 74.2 | 77.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzierżak, R.; Omiotek, Z.; Tkacz, E.; Uhlig, S. Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue. J. Clin. Med. 2022, 11, 4526. https://doi.org/10.3390/jcm11154526
Dzierżak R, Omiotek Z, Tkacz E, Uhlig S. Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue. Journal of Clinical Medicine. 2022; 11(15):4526. https://doi.org/10.3390/jcm11154526
Chicago/Turabian StyleDzierżak, Róża, Zbigniew Omiotek, Ewaryst Tkacz, and Sebastian Uhlig. 2022. "Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue" Journal of Clinical Medicine 11, no. 15: 4526. https://doi.org/10.3390/jcm11154526
APA StyleDzierżak, R., Omiotek, Z., Tkacz, E., & Uhlig, S. (2022). Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue. Journal of Clinical Medicine, 11(15), 4526. https://doi.org/10.3390/jcm11154526








