Machine Learning Can Predict the Probability of Biologic Therapy in Patients with Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Study Population and Outcome
2.3. Potential Predictors and Statistical Analyses
3. Results
3.1. Characteristics of the Study Sample and Incidence of Biologic Therapy
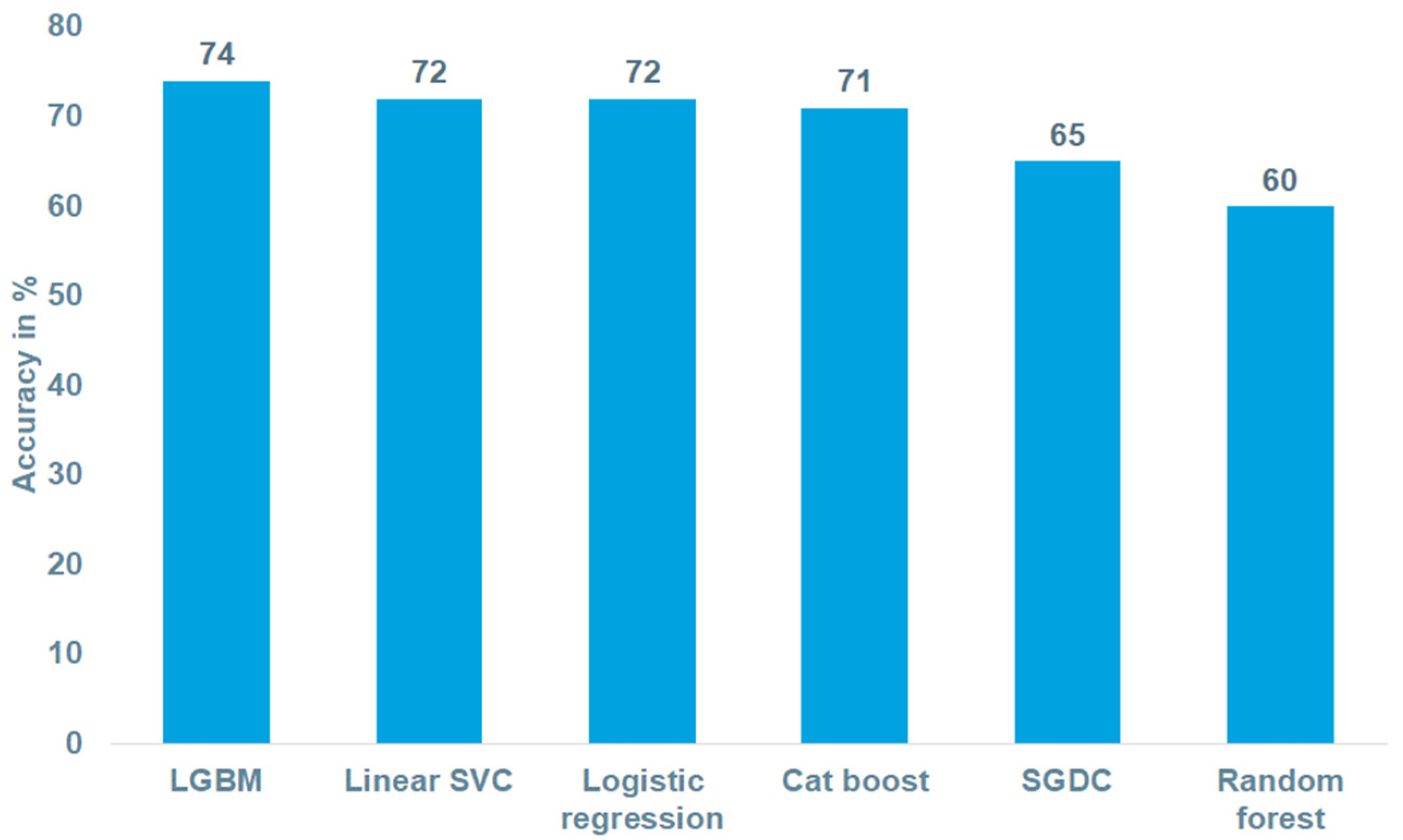
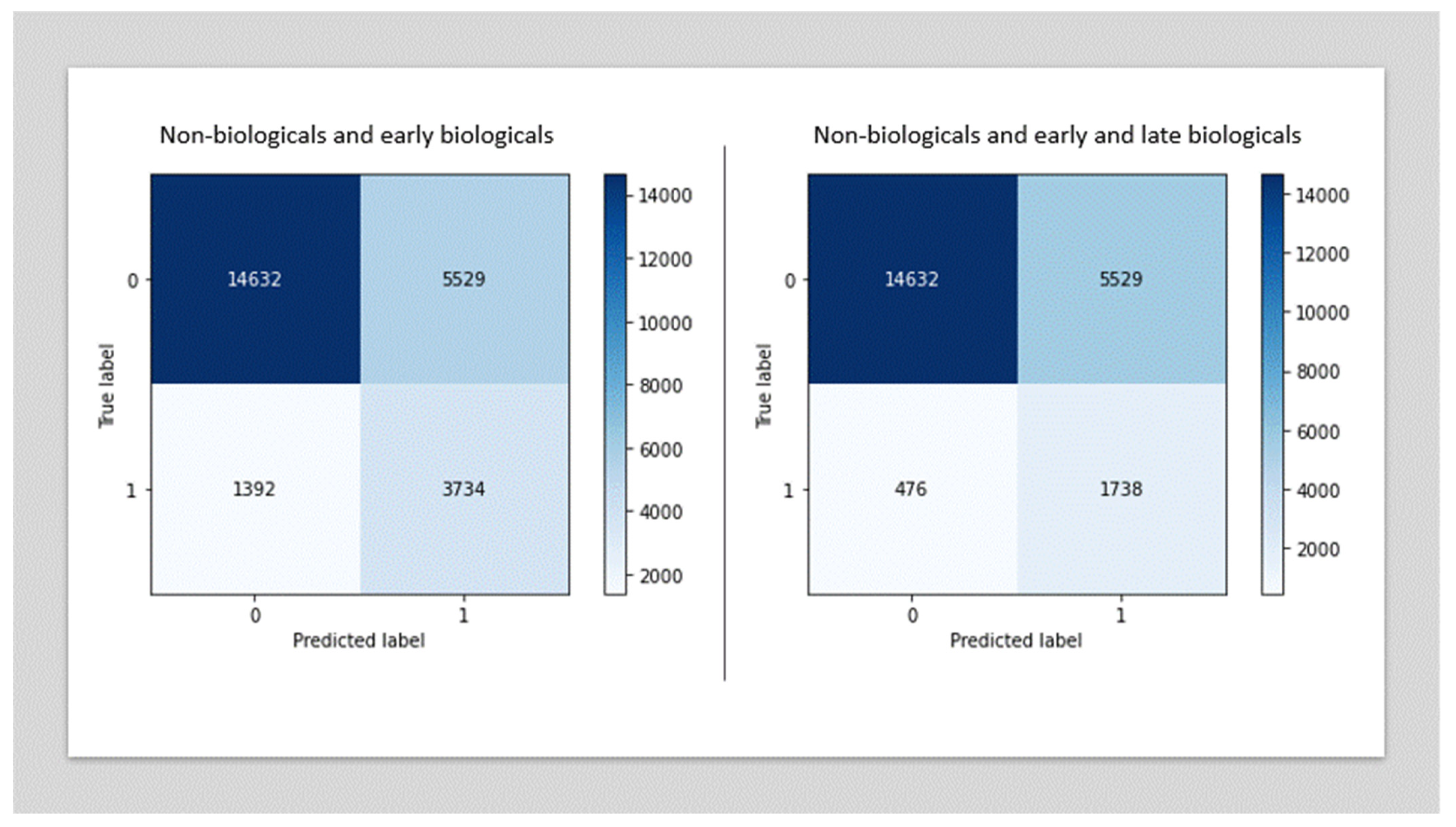
3.2. Performance of the Biologic Therapy Prediction Models
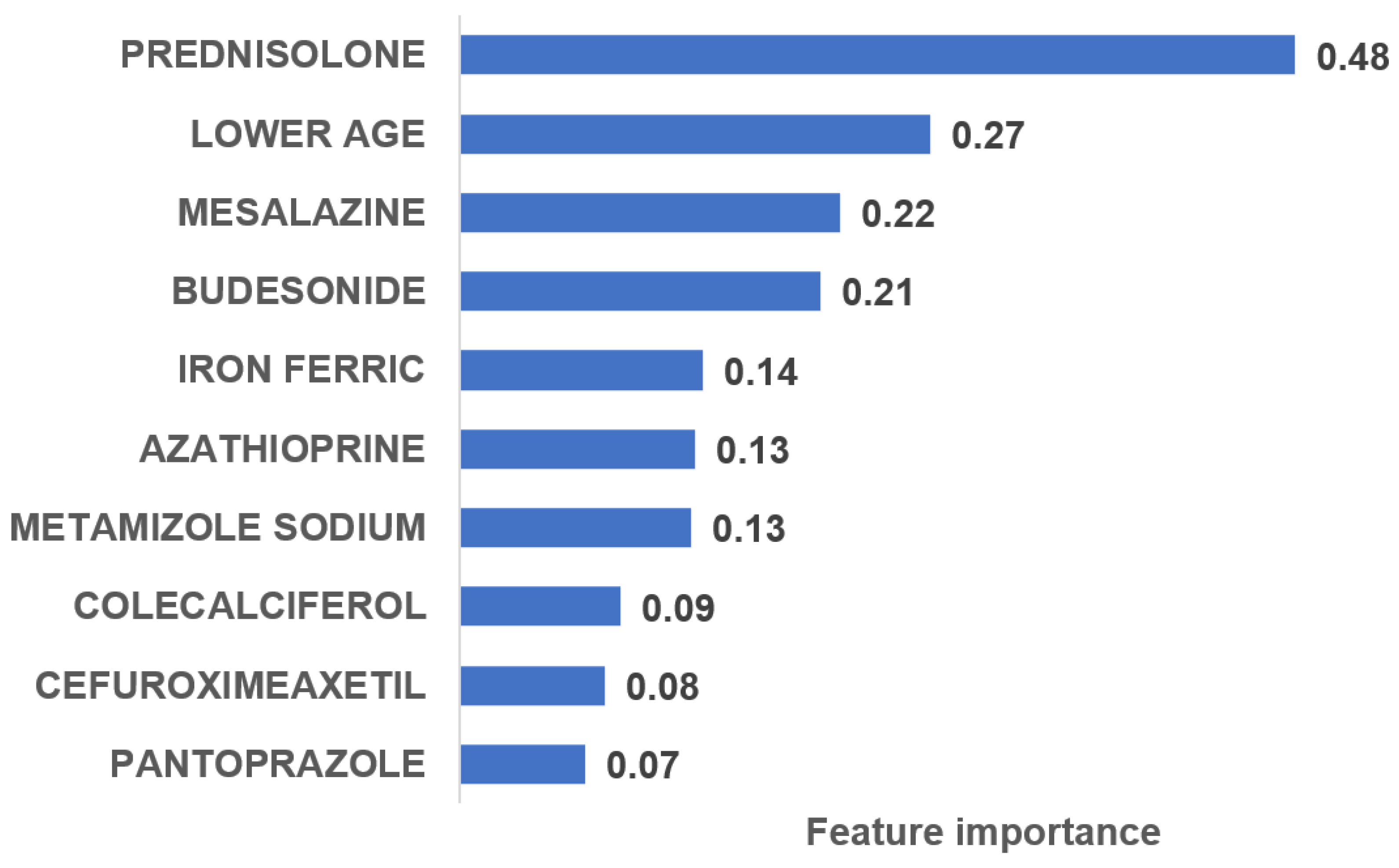
3.3. Most Important Variables Predicting the Probability of Biologic Therapy
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru, K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, N.; Li, M.; He, L.; Xie, B.; Wang, L.; Zhang, R.; Yu, Y.; Sun, X.; Pan, Z.; Wang, K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: A machine learning approach using XGboost. J. Transl. Med. 2020, 18, 462. [Google Scholar] [CrossRef]
- Hae, H.; Kang, S.-J.; Kim, W.-J.; Choi, S.-Y.; Lee, J.-G.; Bae, Y.; Cho, H.; Yang, D.H.; Kang, J.-W.; Lim, T.-H.; et al. Machine learning assessment of myocardial ischemia using angiography: Development and retrospective validation. PLoS Med. 2018, 15, e1002693. [Google Scholar] [CrossRef] [Green Version]
- Richter, H.; Dombrowski, S.; Hamer, H.; Hadji, P.; Kostev, K. Use of a German longitudinal prescription database (LRx) in pharmacoepidemiology. Ger. Med. Sci. 2015, 13, Doc14. [Google Scholar] [CrossRef]
- Helwig, U.; Braegger, F.; Kostev, K.; Schmidt, C. Comparative Analysis of the 3-Year Persistence Rate with Second-Line Vedolizumab and Tumor Necrosis Factor-α Inhibitors in Patients with Inflammatory Bowel Disease Followed in Gastroenterology Practices in Germany. Dig. Dis. 2020, 38, 466–473. [Google Scholar] [CrossRef]
- Rathmann, W.; Bongaerts, B.; Carius, H.-J.; Kruppert, S.; Kostev, K. Basic characteristics and representativeness of the German Disease Analyzer database. Int. J. Clin. Pharmacol. Ther. 2018, 56, 459–466. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A highly efficient gradient boosting decision tree. Adv. Neural Inform. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [Green Version]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Sum, J.; Leung, C.S.; Ho, K. A Limitation of Gradient Descent Learning. IEEE Trans. Neural Networks Learn. Syst. 2020, 31, 2227–2232. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Cosnes, J.; Bourrier, A.; Nion-Larmurier, I.; Sokol, H.; Beaugerie, L.; Seksik, P. Factors affecting outcomes in Crohn’s disease over 15 years. Gut 2012, 61, 1140–1145. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Cheon, J.H.; Moon, C.M.; Park, J.J.; Hong, S.P.; Kim, T.I.; Kim, W.H. Do patients with ulcerative colitis diagnosed at a young age have more severe disease activity than patients diagnosed when older? Digestion 2010, 81, 237–243. [Google Scholar] [CrossRef]
- Choi, Y.I.; Kim, Y.J.; Chung, J.W.; Kim, K.O.; Kim, H.; Park, R.W.; Park, D.K. Effect of age on the initiation of biologic agent therapy in patients with inflammatory bowel disease: Korean common data model cohort study. JMIR Med. Inform. 2020, 8, e15124. [Google Scholar] [CrossRef]
- Sulz, M.C.; Burri, E.; Michetti, P.; Rogler, G.; Peyrin-Biroulet, L.; Seibold, F. Treatment Algorithms for Crohn’s Disease. Digestion 2020, 101, 43–57. [Google Scholar] [CrossRef]
- Kaitha, S.; Bashir, M.; Ali, T. Iron deficiency anemia in inflammatory bowel disease. World J. Gastrointest. Pathophysiol. 2015, 6, 62–72. [Google Scholar] [CrossRef]
- Ham, M.; Longhi, M.S.; Lahiff, C.; Cheifetz, A.; Robson, S.; Moss, A.C. Vitamin D levels in adults with Crohn’s disease are responsive to disease activity and treatment. Inflamm. Bowel Dis. 2014, 20, 856–860. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, O.H.; Hansen, T.I.; Gubatan, J.M.; Jensen, K.B.; Rejnmark, L. Managing Vitamin D deficiency in inflammatory bowel disease. Frontline Gastroenterol. 2019, 10, 394–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeitz, J.; Ak, M.; Muller-Mottet, S.; Scharl, S.; Biedermann, L.; Fournier, N.; Frei, P.; Pittet, V.; Scharl, M.; Fried, M.; et al. Pain in IBD patients: Very frequent and frequently insufficiently taken into account. PLoS ONE 2016, 11, e0156666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, S.; Steinhart, A.H. Antibiotic therapy for Crohn’s disease: A review. Can. J. Gastroenterol. 2006, 20, 651–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.I.; Park, S.J.; Chung, J.W.; Kim, K.O.; Cho, J.H.; Kim, Y.J.; Lee, K.Y.; Kim, K.G.; Park, D.K.; Kim, Y.J. Development of machine learning model to predict the 5-year risk of starting biologic agents in patients with inflammatory bowel disease (Ibd): K-cdm network study. J. Clin. Med. 2020, 9, 3427. [Google Scholar] [CrossRef] [PubMed]
- Olivera, P.; Danese, S.; Jay, N.; Natoli, G.; Peyrin-Biroulet, L. Big data in IBD: A look into the future. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 312–321. [Google Scholar] [CrossRef] [PubMed]
| Therapy Class | Proportion among Biologics Patients in % (n = 15,284) | Proportion among Non-Biologics Patients in % (n = 106,265) | Prevalence Ratio (Biologics/Non-Biologics) | p-Value |
|---|---|---|---|---|
| Calcium | 12.3 | 3.9 | 3.2 | <0.001 |
| Other immunosuppressants * | 33.3 | 11.0 | 3.0 | <0.001 |
| Iron | 24.7 | 8.6 | 2.9 | <0.001 |
| Oral corticosteroids | 71.1 | 26.1 | 2.7 | <0.001 |
| Antidiarrheal microorganisms | 4.7 | 2.0 | 2.4 | <0.001 |
| Intestinal corticosteroids | 64.2 | 33.4 | 1.9 | <0.001 |
| Vitamin D | 20.1 | 10.9 | 1.8 | <0.001 |
| Oral fluoroquinolones | 10.4 | 6.9 | 1.5 | <0.001 |
| Gastroprokinetics | 6.9 | 4.7 | 1.5 | <0.001 |
| Antitussives | 6.8 | 4.7 | 1.4 | <0.001 |
| PPI | 54.0 | 41.4 | 1.3 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schöler, D.; Kostev, K.; Peters, M.; Zamfir, C.; Wolk, A.; Roderburg, C.; Loosen, S.H. Machine Learning Can Predict the Probability of Biologic Therapy in Patients with Inflammatory Bowel Disease. J. Clin. Med. 2022, 11, 4586. https://doi.org/10.3390/jcm11154586
Schöler D, Kostev K, Peters M, Zamfir C, Wolk A, Roderburg C, Loosen SH. Machine Learning Can Predict the Probability of Biologic Therapy in Patients with Inflammatory Bowel Disease. Journal of Clinical Medicine. 2022; 11(15):4586. https://doi.org/10.3390/jcm11154586
Chicago/Turabian StyleSchöler, David, Karel Kostev, Maximilian Peters, Cosmin Zamfir, Agnieszka Wolk, Christoph Roderburg, and Sven H. Loosen. 2022. "Machine Learning Can Predict the Probability of Biologic Therapy in Patients with Inflammatory Bowel Disease" Journal of Clinical Medicine 11, no. 15: 4586. https://doi.org/10.3390/jcm11154586






