Contribution of the Testosterone Androgen Receptor–PARD3B Signaling Axis to Tumorigenesis and Malignance of Glioblastoma Multiforme through Stimulating Cell Proliferation and Colony Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Mining and Analysis
2.2. Bioinformatic Approach
2.3. Cell Culture and Drug Treatment
2.4. Suppression of AR Activity
2.5. Real-Time Polymerase Chain Reaction (PCR)
2.6. Cell Proliferation
2.7. Colony Formation
2.8. Statistical Analysis
3. Results
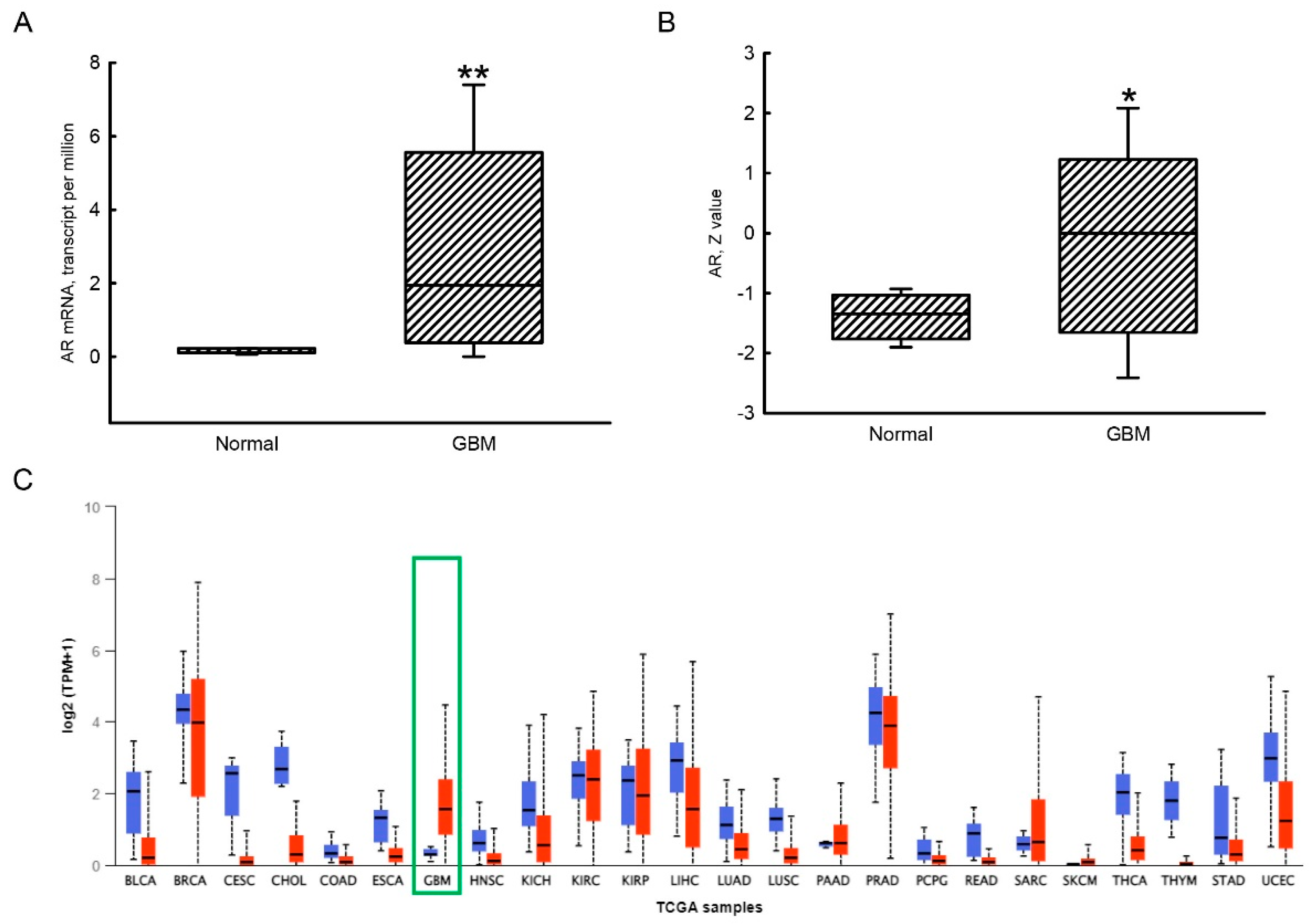
3.1. Upregulation of AR mRNA and Protein Expressions in Human GBM Tissues Compared to Human Normal Brain Tissues
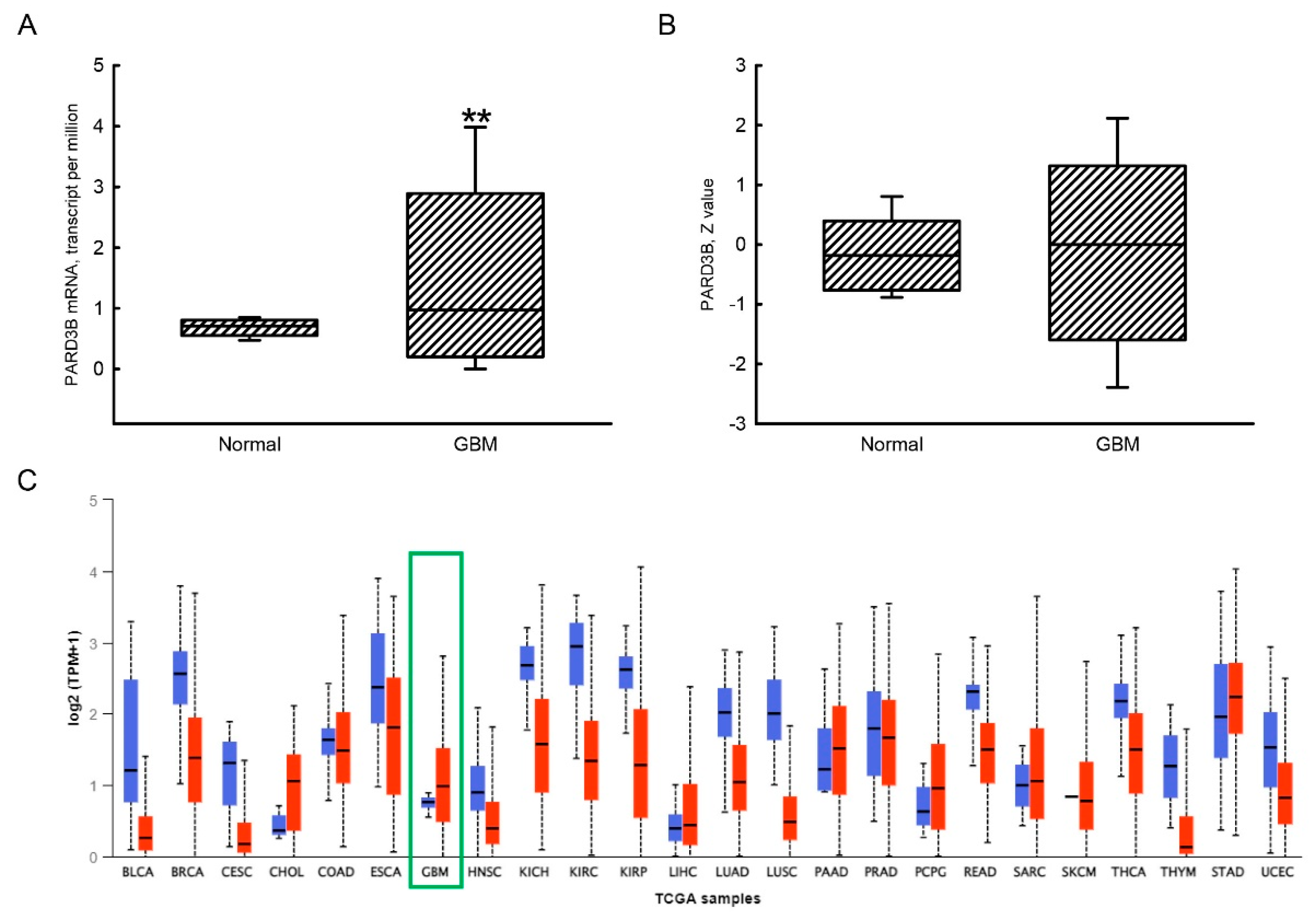
3.2. Increased Expression of the PARD3B Gene in GBM Tissues Compared to Human Normal Brain Tissues
3.3. A Positive Correlation of the AR Gene Expression with PARD3B Expression in Human GBM Tissues
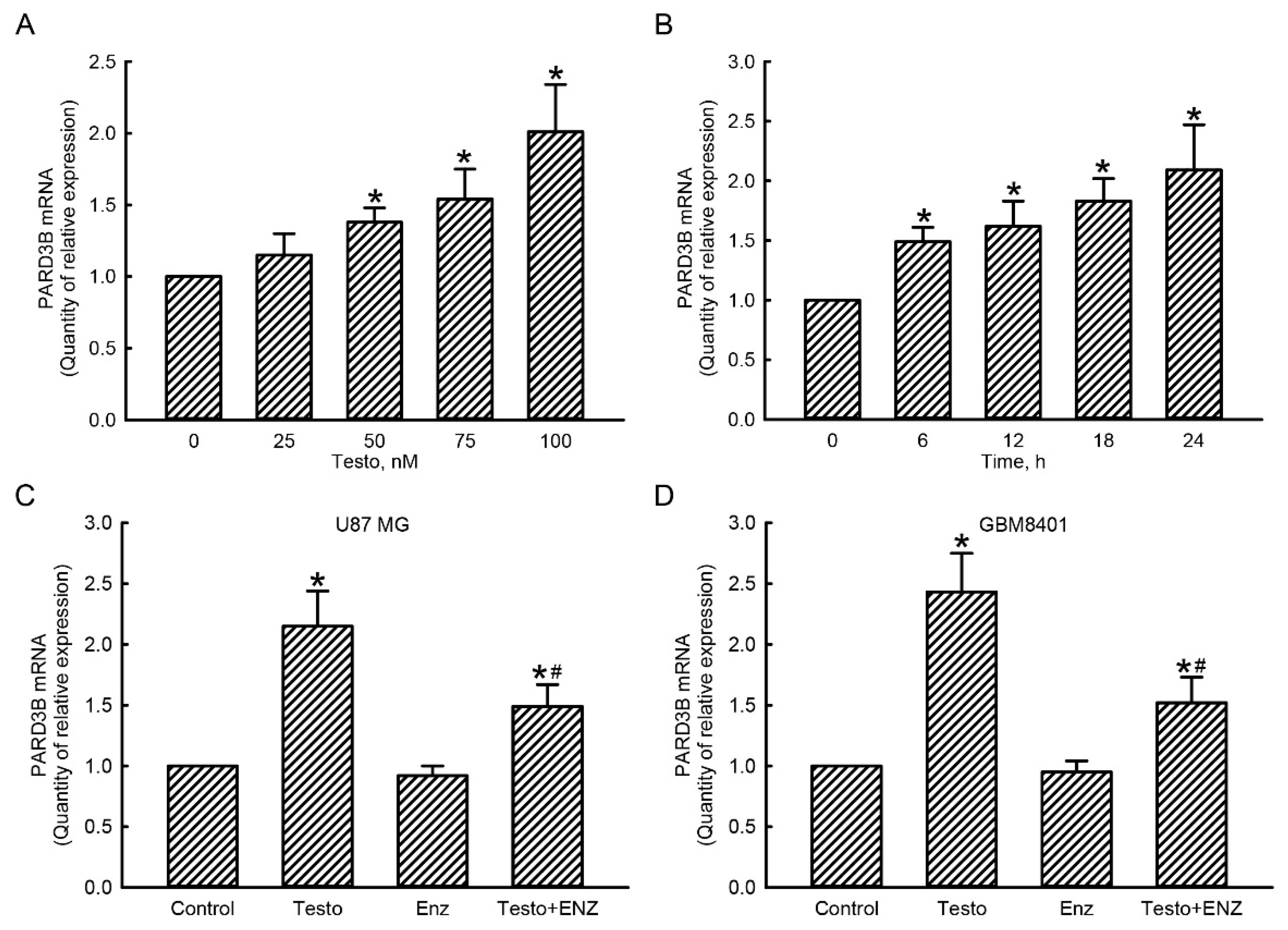
3.4. The Testosterone AR Signaling Axis Was Involved in Regulation of PARD3B Gene Expression in Human Glioblastoma Cells
3.5. The Testosterone AR Signaling Axis Contributes to Proliferation of Human Glioblastoma Cells
3.6. The Testosterone AR Signaling Axis Contributes to Regulation of Proliferation of Human Glioblastoma Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yao, M.; Li, S.; Wu, X.; Diao, S.; Zhang, G.; He, H.; Bian, L.; Lu, Y. Cellular origin of glioblastoma and its implication in precision therapy. Cell. Mol. Immunol. 2018, 15, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, A.; Rossato, M.; Lombardi, G.; Benfatto, S.; Lavezzari, D.; de Salvo, G.L.; Indraccolo, S.; Dechecchi, M.C.; Prandini, P.; Gambari, R.; et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro-Oncol. 2021, 23, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Shahcheraghi, S.H.; Tchokonte-Nana, V.; Lotfi, M.; Lotfi, M.; Ghorbani, A.; Sadeghnia, H.R. Wnt/beta-catenin and PI3K/Akt/mTOR signaling pathways in glioblastoma: Two main targets for drug design: A review. Curr. Pharm. Des. 2020, 26, 1729–1741. [Google Scholar] [CrossRef]
- Litak, J.; Mazurek, M.; Grochowski, C.; Kamieniak, P.; Roliński, J. PD-L1/PD-1 Axis in Glioblastoma Multiforme. Int. J. Mol. Sci. 2019, 20, 5347. [Google Scholar] [CrossRef]
- Roche, P.J.; Hoare, S.A.; Parker, M.G. A consensus DNA-binding site for the androgen receptor. Mol. Endocrinol. 1992, 6, 2229–2235. [Google Scholar]
- Tsai, M.J.; O’Malley, B.W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994, 63, 451–486. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, M.; Shen, C.; Liu, G.; Zhang, F.; Hou, J.; Yao, W. LncRNA PCBP1-AS1-mediated AR/AR-V7 deubiquitination enhances prostate cancer enzalutamide resistance. Cell Death Dis. 2021, 12, 856. [Google Scholar] [CrossRef]
- Jamroze, A.; Chatta, G.; Tang, D.G. Androgen receptor (AR) heterogeneity in prostate cancer and therapy resistance. Cancer Lett. 2021, 518, 1–9. [Google Scholar] [CrossRef]
- Łysiak, M.; Trybuła, M.; Mudaisi, M.; Bratthäll, C.; Strandeus, M.; Milos, P.; Hallbeck, M.; Malmström, A. The sex-dependent role of the androgen receptor in glioblastoma: Results of molecular analyses. Mol. Oncol. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Carrano, A.; Juarez, J.J.; Incontri, D.; Ibarra, A.; Guerrero Cazares, H. Sex-specific differences in glioblastoma. Cells 2021, 10, 1783. [Google Scholar] [CrossRef]
- Wei, H.; Wen, W. Phase separation in cell polarity. Biochemistry 2021, 60, 2677–2684. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, A.; Näthke, I. Cell polarity in development and cancer. Nat. Cell Biol. 2007, 9, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.J. Par-3 family proteins in cell polarity & adhesion. FEBS J. 2022, 289, 596–613. [Google Scholar] [PubMed]
- Atashrazm, F.; Ellis, S. The polarity protein PARD3 and cancer. Oncogene 2021, 40, 4245–4262. [Google Scholar] [CrossRef]
- Kohjima, M.; Noda, Y.; Takeya, R.; Saito, N.; Takeuchi, K.; Sumimoto, H. PAR3beta, a novel homologue of the cell polarity protein PAR3, localizes to tight junctions. Biochem. Biophys. Res. Commun. 2002, 299, 641–646. [Google Scholar] [CrossRef]
- Li, T.; Liu, D.; Lei, X.; Jiang, Q. Par3L enhances colorectal cancer cell survival by inhibiting Lkb1/AMPK signaling pathway. Biochem. Biophys. Res. Commun. 2017, 482, 1037–1041. [Google Scholar] [CrossRef]
- Ko, K.; Kitani, T.; Harris, B.T.; Anaizi, A.N.; Solomon, D.; Perry, A.; Toretsky, J.; Ozdemirli, M. A novel PARD3B-NUTM1 fusion in an aggressive primary CNS embryonal tumor in a young adult. Acta Neuropathol. Commun. 2020, 8, 112. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Sun, D.P.; Lee, Y.W.; Chen, J.T.; Lin, Y.W.; Chen, R.M. The bradykinin-BDKRB1 axis regulates aquaporin 4 gene expression and consequential migration and invasion of malignant glioblastoma cells via a Ca2+-MEK1-ERK1/2-NF-κB Mechanism. Cancers 2020, 12, 667. [Google Scholar] [CrossRef]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG glioma cell line: Good news and bad news. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Yeh, M.Y.; Tu, Y.C.; Han, S.H.; Wang, Y.C. Establishment and characterization of a malignant glioma cell line, GBM8401/TSGH, NDMC. J. Surg. Oncol. 1988, 38, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.W.; Chen, J.T.; Hong, C.Y.; Lin, Y.L.; Wang, K.T.; Yao, C.J.; Lai, G.M.; Chen, R.M. Honokiol traverses the blood-brain barrier and induces apoptosis of neuroblastoma cells via an intrinsic Bax-mitochondrion-cytochrome c-caspase protease pathway. Neuro-Oncol. 2012, 14, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, R.; Sawant, M.; Renganathan, A.; Mahajan, K.; Kim, E.H.; Luo, J.; Dang, H.X.; Maher, C.A.; Feng, F.Y.; Mahajan, N.P. Loss of long noncoding RNA NXTAR in prostate cancer augments androgen receptor expression and enzalutamide resistance. Cancer Res. 2022, 82, 155–168. [Google Scholar] [CrossRef]
- Wu, G.J.; Chen, K.Y.; Yang, J.D.; Liu, S.H.; Chen, R.M. Naringin improves osteoblast mineralization and bone healing and strength through estrogen receptor alpha-dependent regulation of alkaline phosphatase gene expression. J. Agric. Food Chem. 2021, 69, 13020–13033. [Google Scholar] [CrossRef]
- Stubbs, A.P.; Engelman, J.L.; Walker, J.I.H.; Faik, P.; Murphy, G.M.; Wilkinson, M.L. Measurement of androgen receptor expression in adult liver, fetal liver, and Hep-G2 cells by the polymerase chain reaction. Gut 1994, 35, 683–686. [Google Scholar] [CrossRef]
- Joo, K.M.; Kim, S.; Koo, Y.J.; Lee, M.; Lee, S.H.; Choi, D.; Lim, K.M. Development and validation of UPLC method for WST-1 cell viability assay and its application to MCTT HCE™ eye irritation test for colorful substances. Toxicol. In Vitro 2019, 60, 412–419. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro-Oncol. 2019, 21 (Suppl. 5), v1–v100. [Google Scholar] [CrossRef]
- Shi, L.; Yan, Y.; He, Y.; Yan, B.; Pan, Y.; Orme, J.J.; Zhang, J.; Xu, W.; Pang, J.; Huang, H. Mutated SPOP E3 ligase promotes 17βHSD4 protein degradation to drive androgenesis and prostate cancer progression. Cancer Res. 2021, 81, 3593–3606. [Google Scholar] [CrossRef]
- Rajendran, V.; Jain, M.V. In vitro tumorigenic assay: Colony forming assay for cancer stem cells. Methods Mol. Biol. 2018, 1692, 89–95. [Google Scholar]
- Chen, X.; Zeh, H.J.; Kang, R.; Kroemer, G.; Tang, D. Cell death in pancreatic cancer: From pathogenesis to therapy. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 804–823. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, T.; Chen, Y.; Bao, J.; Wu, D.; Hu, X.; Feng, C.; Xu, L.; Li, M.; Li, G.; et al. USP5 sustains the proliferation of glioblastoma through stabilization of cyclinD1. Front. Pharmacol. 2021, 12, 720307. [Google Scholar] [CrossRef] [PubMed]
- Deldar Abad Paskeh, M.; Asadi, S.; Zabolian, A.; Saleki, H.; Khoshbakht, M.A.; Sabet, S.; Naghdi, M.J.; Hashemi, M.; Hushmandi, K.; Ashrafizadeh, M.; et al. Targeting cancer stem cells by dietary agents: An important therapeutic strategy against human malignancies. Int. J. Mol. Sci. 2021, 22, 11669. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.-D.; Chen, J.-T.; Liu, S.-H.; Chen, R.-M. Contribution of the Testosterone Androgen Receptor–PARD3B Signaling Axis to Tumorigenesis and Malignance of Glioblastoma Multiforme through Stimulating Cell Proliferation and Colony Formation. J. Clin. Med. 2022, 11, 4818. https://doi.org/10.3390/jcm11164818
Yang J-D, Chen J-T, Liu S-H, Chen R-M. Contribution of the Testosterone Androgen Receptor–PARD3B Signaling Axis to Tumorigenesis and Malignance of Glioblastoma Multiforme through Stimulating Cell Proliferation and Colony Formation. Journal of Clinical Medicine. 2022; 11(16):4818. https://doi.org/10.3390/jcm11164818
Chicago/Turabian StyleYang, Jr-Di, Jui-Tai Chen, Shing-Hwa Liu, and Ruei-Ming Chen. 2022. "Contribution of the Testosterone Androgen Receptor–PARD3B Signaling Axis to Tumorigenesis and Malignance of Glioblastoma Multiforme through Stimulating Cell Proliferation and Colony Formation" Journal of Clinical Medicine 11, no. 16: 4818. https://doi.org/10.3390/jcm11164818




