Artificial Intelligence-Assisted Renal Pathology: Advances and Prospects
Abstract
:1. Background
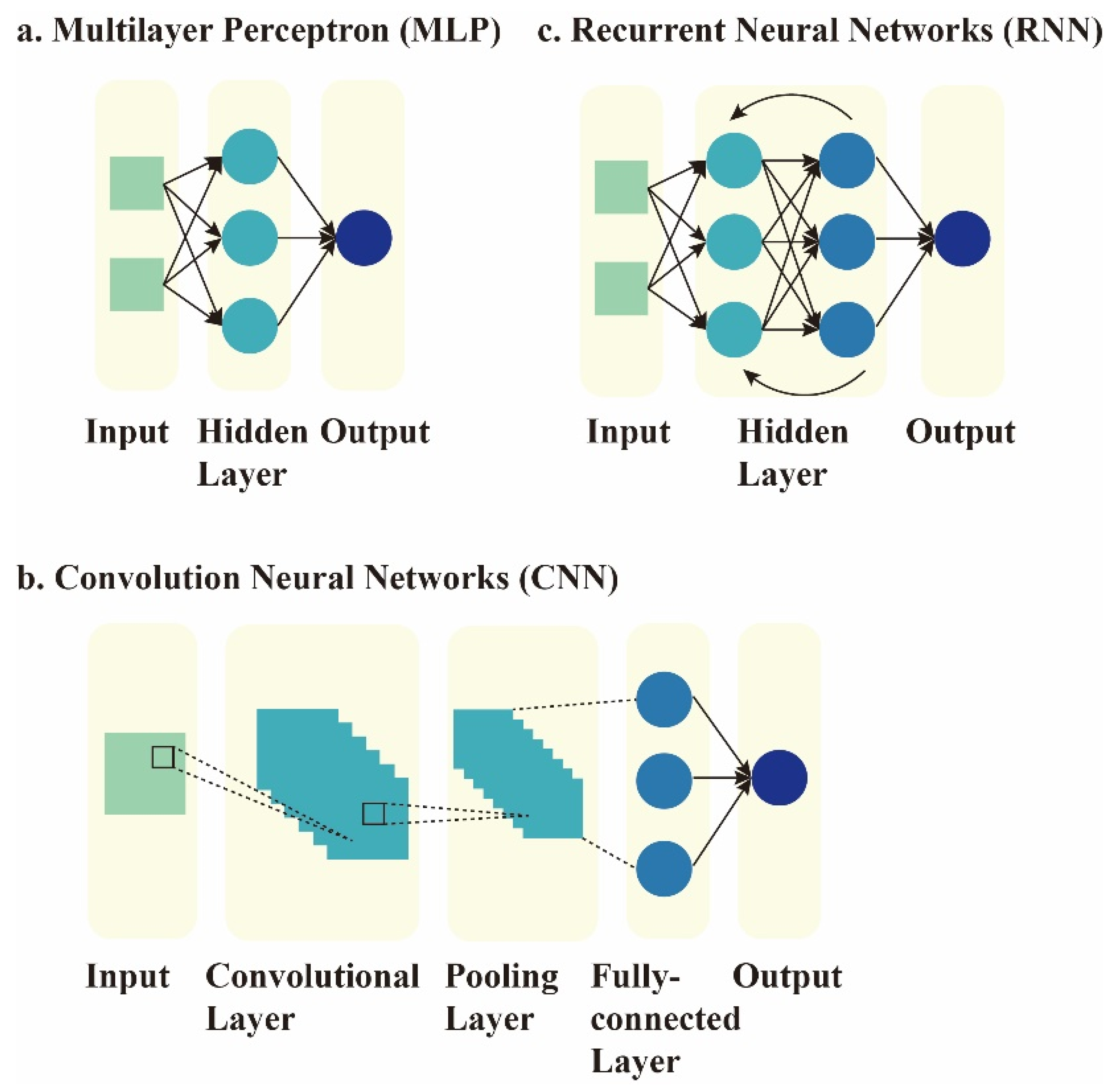
2. Current Concepts of AI
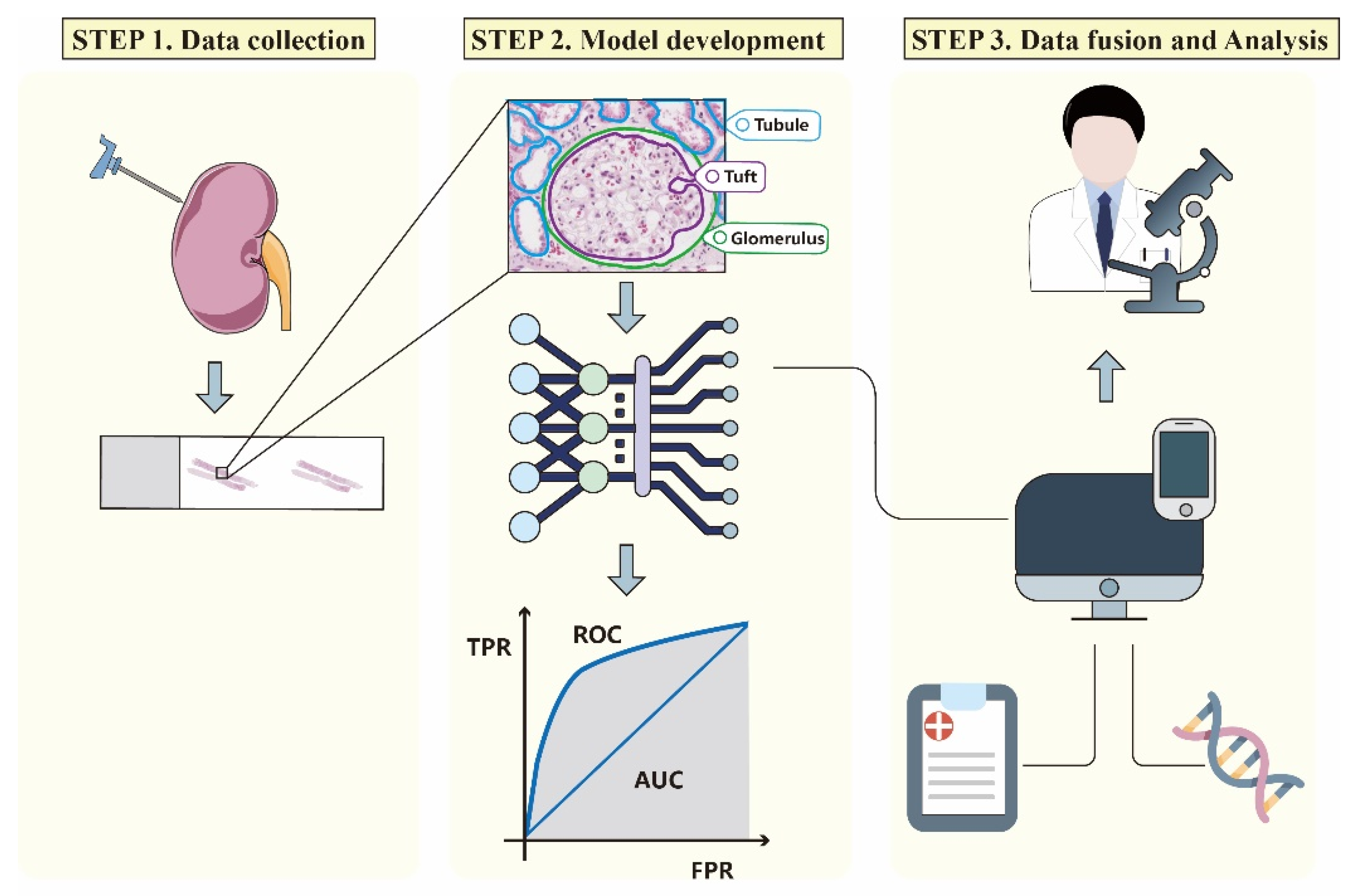
3. AI Image Processing Workflow
3.1. Data Selection, Collection, and Annotation
3.2. Image Pre-Processing and Model Development
3.3. Model Verification and Data Fusion
4. Application of AI in Nephropathology
4.1. Detection and Segmentation of Kidney Structures
| Object | Author | Year | Task | Methods | Slides | Main Results | Ref. |
|---|---|---|---|---|---|---|---|
| Normal glomeruli | Simon et al. | 2018 | Localization of glomeruli | CNN, SVM | 15 WSIs, healthy mice (H&E) 15 WSIs, STZ-mice (H&E) 15 WSIs, rat (CR, H&E, Jones, PAS, and Gömorri trichrome) 25 WSIs, DN patients (PAS) | Glomerular detection in mouse: precision: >90%; recall: >70% | [36] |
| Bukowy et al. | 2018 | Localization of glomeruli with trichrome-staining | Alexnet CNN | 87 WSIs, rat (Gömöri or Masson trichrome) | Average precision: 96.94%; recall: 96.79% | [52] | |
| Sheehan et al. | 2018 | Segmentation and quantification of glomeruli | Ilastik | 738 images, mice (PAS) | Precision: 98.4%; recall: 95.2%, F-score: 96.0% | [53] | |
| Wilbur et al. | 2021 | Detection of glomeruli of four different stains across institutions | CNN | 284 WSIs, human (H&E, PAS, PASM, trichrome) | Sensitivity: intra-institutional: 90–93%; interinstitutional: 77%; combined: 86% Modified specificity: intra-institutional: 86–98%; interinstitutional: 97%; combined: 92% | [54,55] | |
| Proliferative glomeruli | Chagas et al. | 2020 | Binary or multiple classification of hypercellularity | CNN, SVM | 811 images, human (H&E, PAS) | Binary classification: average accuracy: nearly 100% Multiple classification: average accuracy: 82% | [39] |
| Barros et al. | 2017 | Segmentation and classification of glomeruli w/ or w/o proliferative changes | kNN | 811 images, human (H&E, PAS) | Generalization set: precision: 92.3%; recall: 88.0%; accuracy: 88% | [56] | |
| Sclerotic glomeruli | Kannan et al. | 2019 | Classification of normal and sclerosed glomeruli | Inception v3 CNN | 171 WSIs, human (trichrome) | Accuracy: 92.67% ± 2.02%; kappa: 0.8681 ± 0.0392 | [34] |
| Jiang et al. | 2021 | Detection, classification, and segmentation of glomeruli into three categories | Cascade mask region-based CNN | 1123 snapshots, human (H&E, PAS, PASM, Masson) 348 WSIs, human (H&E, PAS, PASM, Masson) | Snapshot group: F1-score: total glomeruli, GN, global sclerosis, and glomerular with other lesions (0.914, 0.896, 0.681, 0.756) WSI group: F1-score: total glomeruli, GN, global sclerosis, and glomerular with other lesions (0.940, 0.839, 0.806, 0.753) | [43] | |
| Lutnick et al. | 2020 | Label-free classification of glomeruli by Tervaert class and the presence of sclerosis | VAE-GAN | 1193 individual glomeruli (H&E, PAS) 121 WSIs, human (PAS) | Cohen’s kappa values: Tervaert class: 0.87 sclerosis: 0.78 | [44] | |
| Lu et al. | 2022 | Quantification and subtype classification of global glomerulosclerosis | Transfer learning | 7841 globally sclerotic glomeruli of three distinct categories | Pretrained dataset: F1-score: 0.778 External dataset: AUC: 0.994 | [45] | |
| Bueno et al. | 2020 | Semantic and classification of normal and sclerosed glomeruli | SegNet-VGG19+ AlexNet CNN | 47 WSIs, human (PAS) | Accuracy: 98.16% F1-score: 0.994 | [57] | |
| Gallego et al. | 2021 | Classification of normal and sclerosed glomeruli | U-Net CNN | 51 WSIs, human (PAS, H&E) | F1-score PAS: normal glomeruli: 97.5%; sclerosed glomeruli: 68.8% H&E: normal glomeruli: 90.8%; sclerosed glomeruli: 78.1% Average: normal glomeruli: 94.5%; sclerosed glomeruli: 76.8% | [58] | |
| Francesco et al. | 2022 | Classification of sclerotic and non-sclerotic glomeruli | IBM Watson | 26 WSIs, human (PAS) | Mean accuracy: 99% | [59,60,61] | |
| Marsh et al. | 2018 | Classification of non-sclerosed and sclerosed glomeruli | VGG16 CNN | 48 WSIs, human (frozen sections: H&E) | Non-sclerosed glomeruli: precision: 81.3%; recall: 88.5%; F1-Score: 84.8% Sclerosed glomeruli: precision: 60.7%; recall: 69.8%; F1-score: 64.9% | [62] | |
| Li et al. | 2021 | Quantification of non-sclerotic and sclerotic glomeruli | U-Net CNN | 258 WSIs, human (frozen sections) | Non-sclerosed glomeruli: Dice similarity coefficient: 0.90; recall: 0.90; F1-score: 0.93; precision: 0.96 Sclerosed glomeruli: Dice similarity coefficient: 0.93; recall: 0.87; F1-score: 0.96; precision: 0.81 | [63] | |
| Marsh et al. | 2021 | Quantification of percent global glomerulosclerosis | VGG16 CNN | 149 WSIs, human (frozen and permanent sections: H&E) | Higher correlation with annotations (r = 0.916; 95% CI, 0.886–0.939) than on-call pathologists (r = 0.884; 95% CI, 0.825–0.923) Lower model prediction error for single levels (RMSE, 5.631; 95% CI, 4.735–6.517) than on-call pathologists (RMSE, 6.523; 95% CI, 5.191–7.783) Decreased the likelihood of unnecessary organ discard by 37% compared with pathologists | [64] | |
| Glomeruli with multiple pathological changes | Weis et al. | 2022 | Classification of 9 glomerular structural changes | CNN | 23,395 glomerular images, human (PAS) | Kappa-values: 0.838–0.938 | [46] |
| Yamaguchi et al. | 2021 | Classification of glomerular images of 12 features | ResNet50 CNN | 293 WSIs, human (PAS) | ROC–AUC: 0.65–0.98. (“capillary collapse”: 0.98) | [47] | |
| Zhang et al. | 2022 | Segmentation of glomeruli and classification of the deposition pattern in immunofluorescence image | U-Net, MANet | 4779 images, human (IF) | Deposition region: accuracy: 98% Deposition appearance: accuracy: 95% Label fusion: accuracy: >90% | [48] | |
| Uchino et al. | 2020 | Classification of glomeruli of 7 pathological changes | InceptionV3 CNN | 283 WSIs, human (PAS, PASM) | Global sclerosis: AUC: PAS: 0.986; PASM: 0.983 Other pathological findings: AUC: 0.59–0.87 (close to those of nephrologists) | [65] | |
| Yang et al. | 2021 | Detection, classification, lesion identification of glomerular disease | Mask R-CNN, LSTM RNN, ResNeXt-101 | Detection: 1379 slides, human (H&E, PAS, TRI, PAM) Classification: 653 cases, human | Detection: F1-scores: up to 0.944 Classification: accuracies: up to 0.940 Lesion identification: AUC: up to 0.947 | [66] | |
| Nan et al. | 2022 | Classification of five subcategories of IgAN glomerular lesions | UAAN | 400 WSIs, human (PAS) | Accuracy: 93.0% Fl-score: 92.9% | [67] | |
| Other kidney structures | Hermsen et al. | 2019 | Multiclass segmentation of kidney biopsies | U-Net CNN | 132 WSIs, human (PAS) | Weighted mean Dice coefficients of all classes: 0.80–0.84 Mean intraclass correlation coefficient (pathologists versus the network): 0.94 | [31] |
| Sheehan et al. | 2019 | Identification of histological differences between mice of different genotypes according to segmentation of kidney structure | AlexNet DNN, SVM | 90 WSIs, mice (PAS) | Identification of previously neglected histologic features, including vacuoles, nuclear count, and proximal tubule brush border integrity, to distinguish mice of different genotypes | [68] | |
| Bouteldja et al. | 2021 | Segmentation of kidney tissue | U-Net CNN | 168 WSIs, healthy and diseased mouse, pig, marmoset, bear and rat, human (PAS) | Multiclass segmentation performance was very high in all murine disease models (Dice score: 73.5–98.8) and in other species (Dice score: 76.6–99) | [69] | |
| Jayapandian et al. | 2021 | Segmentation of histologic structures in multi-stained kidney biopsies | U-Net CNN | 459 WSIs, human (H&E, PAS, TRI, SIL) | F-scores: PAS (optimal): glomerular tufts: 0.93; glomerular tuft plus Bowman’s capsule: 0.94; proximal tubules: 0.91; distal tubular segments: 0.93; peritubular capillaries: 0.81; arteries and afferent arterioles: 0.85 | [70] | |
| Govind et al. | 2021 | Label-free identification and quantification of podocyte | Cloud-based AI | 122 WSIs, mouse, rat, and human (PAS) | Sensitivity/specificity: mouse: 0.80/0.80; rat: 0.81/0.86; human: 0.80/0.91 | [71] | |
| Renal cell carcinoma | Michael Fenstermaker et al. | 2020 | Identification and evaluation of renal cell carcinoma | CNN | 12,168 RCC samples, human | Accuracy: normal parenchyma vs. RCC: 99.1%; clear cell, papillary, and chromophobe histiotypes: 97.5%; Fuhrman grade: 98.4% | [51] |
| Eliana Marostica et al. | 2021 | Classification and prediction of clinical outcomes in subtypes of renal cell carcinoma | Deep convolutional neural networks (DCNN) | 231 slides (chRCC), 1657 slides (ccRCC), 475 slides (pRCC), human | AUC: detection of malignancy: 0.964–0.985; diagnosis of RCC histologic subtypes: 0.953–0.993 | [72] | |
| Sairam Tabibu et al. | 2019 | Classification and survival prediction of renal cell carcinoma | CNN | 1027 images (ccRCC), 303 images (pRCC), and 254 images (chRCC), human | Classification of RCC histologic subtypes: 94.07% | [73] | |
| Mengdan Zhu et al. | 2021 | Classification of 4 subtypes of renal cell carcinoma | Deep neural network | 1074 WSIs, human | AUC: 0.97–0.98 | [74] |
4.2. Auxiliary Diagnosis of Renal Pathological Changes
4.2.1. Renal Interstitial Fibrosis
4.2.2. Lupus Nephritis
4.2.3. Diabetic Nephropathy
4.2.4. IgA Nephropathy
| Disease | Author | Year | Task | Methods | Slides | Main Results | Ref. |
|---|---|---|---|---|---|---|---|
| Renal interstitial fibrosis | Ginley et al. | 2021 | Detection and quantification of IFTA and glomerulosclerosis | CNN | 116 WSIs, human (PAS) | High levels of agreement between CNN and four renal pathologists: IFTA agreement: ICC: 0.97 (0.94–0.99) glomerulosclerosis agreement: ICC: 0.91 (0.84–0.96) | [81] |
| Marechal et al. | 2022 | Automated segmentation of kidney tissue | CNN | 241 samples of healthy kidney tissue, human | AUC: tubular atrophy: 0.92 interstitial fibrosis level: 0.91 vascular luminal stenosis (>50%): 0.85 | [82] | |
| Z. Yi et al. | 2022 | Recognition of interstitial fibrosis, tubular atrophy, and mononuclear leukocyte infiltration | U-Net and mask R-CNN algorithms | 789 transplant biopsies, human (PAS) | Recognition of abnormal tubules: TPR: 84% | [83] | |
| Farris et al. | 2021 | Quantification of interstitial fibrosis | VGG19 CNN | 100 biopsy specimens, human | Moderate agreement between algorithm and pathologists: correlation coefficient: 0.46 (0.40–0.52) | [109] | |
| Lupus nephritis | Yang et al. | 2021 | Identification of glomerular lesion | ResNeXt-101 | 146 class III or IV (±class V) lupus nephritis biopsies, human (H&E) | Identification of globally sclerotic glomeruli: accuracy: 0.98–0.99 AUC of each kind of lesion: 0.687–0.946 | [66] |
| Zheng et al. | 2021 | Classification of glomerular pathological findings in LN | YOLOv4 and VGG16 | 349 annotated WSIs (PAS) 321 unannotated WSIs (PAS) | Glomerular level: F1 (“slight” and “severe”): 0.924–0.952 Per-patient kidney level: weighted kappa with nephropathologist: 0.855 | [88] | |
| Pan et al. | 2021 | Classification of kidney diseases in IF images | AlexNet | 655 IF images of IgAN (IF) | AUC of non-blurred IF images: 0.997 AUC of blurred IF images: 0.992 | [90] | |
| Cicalese et al. | 2021 | Classification of LGN | Uncertainty-guided Bayesian classification scheme | 87 biopsy specimens, mice (PAS) | Weighted glomerular-level accuracy: 94.5%, weighted kidney-level accuracy: 96.6% | [110] | |
| Diabetic nephropathy | Ginley B et al. | 2019 | Classification of glomerular lesions | CNN | 54 WSIs, human (PAS); 24 WSIs, mice (PAS) | Moderate Cohen’s kappa κ of agreement with a senior pathologist: 0.55 (0.40–0.60) | [96] |
| Kitamura S et al. | 2020 | Diagnosis of diabetic nephropathy with renal pathological immunofluorescence | Deep learning | 885 renal immunofluorescent images, human | Six programs showed 100% accuracy, precision, and recall, and the AUC was 1.000 | [97] | |
| Hacking S et al. | 2021 | Classification of medical kidney disease on electron microscopy images | MedKidneyEM-v1 classifier (deep learning) | 600 images | Diabetic glomerulosclerosis: precision: 88.89% recall: 66.67% | [98] | |
| Ravi et al. | 2019 | Detection of glomerulosclerosis in DN | Genetic k-means | - | Detect 99% of pathological DN glomerulosclerosis | [111] | |
| IgA nephropathy | Zeng et al. | 2020 | Identification of glomerular lesions and intrinsic glomerular cell types | ARPS | 400 WSIs, human (PAS) | Evaluation of global, segmental glomerular sclerosis, and crescents: Cohen’s kappa values: 1.0, 0.776, 0.861 | [106] |
| Sato N et al. | 2021 | Evaluation of the relationship between kidney histological images and clinical information | CNN | 68 WSIs, human (H&E) | Significant relationship between the score of the patch-based cluster containing crescentic glomeruli and SCr: coefficient = 0.09, p = 0.019 | [107] | |
| Purwar R et al. | 2022 | Detection of mesangial hypercellularity of MEST-C score | CNN | 138 individual glomerulus images of IgA patients | Accuracy: 90 ± 2%, sensitivity: 90.4%, specificity: 80% | [112] |
4.3. Prognosis Prediction
| Author | Year | Task | Methods | Slides | Main results | Ref. |
|---|---|---|---|---|---|---|
| Kolachalama et al. | 2018 | Prediction of the 1-, 3-, and 5-year renal survival rates | CNN | 300 biopsies, human (trichrome-stain) | AUC of 1-, 3-, and 5-year renal survival: 0.878, 0.875, and 0.904 | [41] |
| Lee et al. | 2022 | Prediction of the baseline eGFR and 1-year change | ML | 161 biopsies human (trichrome-stain) | AUC of baseline eGFR: 0.93, AUC of 1-year eGFR: 0.80 | [113] |
| Ledbetter et al. | 2017 | Prediction of 1-year eGFR | CNN | 80 biopsies, human (trichrome-stain, PAS) | Mean absolute error of 17.55 mL/min | [114] |
5. Challenges and Limitations
5.1. Lack of Accountability
5.2. Insufficient Data
5.3. Variations of the Image Quality
6. Outlook for the Future
6.1. Fusion of Data
6.2. Application of State-of-the-Art Technology
6.3. Make Full Use of the Unknown
6.4. Association of AI with Nephrologists
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, C.; Hwang, M.; Huang, T.H.; Chen, Y.M.J.; Chang, Y.J.; Ho, T.H.; Huang, J.; Hwang, K.S.; Ho, W.H. Application of artificial intelligence ensemble learning model in early prediction of atrial fibrillation. BMC Bioinform. 2021, 22, 93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F. Application of machine learning in CT images and X-rays of COVID-19 pneumonia. Medicine 2021, 100, e26855. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Zhou, G.M.; Li, W.G. Application of artificial intelligence in renal pathological diagnosis. Chin. J. Kidney Dis. Investig. 2020, 9, 135–137. [Google Scholar]
- Ahuja, A.S. The impact of artificial intelligence in medicine on the future role of the physician. PeerJ 2019, 7, e7702. [Google Scholar] [CrossRef] [PubMed]
- Woolf, B.P. Chapter 7—Machine Learning. In Building Intelligent Interactive Tutors; Woolf, B.P., Ed.; Morgan Kaufmann: San Francisco, CA, USA, 2009; pp. 221–297. [Google Scholar]
- Doshi, M.; Varghese, A. Chapter 12—Smart agriculture using renewable energy and AI-powered IoT. In AI, Edge and IoT-Based Smart Agriculture; Abraham, A., Dash, S., Rodrigues, J.J.P.C., Acharya, B., Pani, S.K., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 205–225. [Google Scholar]
- Connor, C.W. Artificial Intelligence and Machine Learning in Anesthesiology. Anesthesiology 2019, 131, 1346–1359. [Google Scholar] [CrossRef] [Green Version]
- Currie, G.; Hawk, K.E.; Rohren, E.; Vial, A.; Klein, R. Machine Learning and Deep Learning in Medical Imaging: Intelligent Imaging. J. Med. Imaging Radiat Sci. 2019, 50, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Mao, S.; Wang, B.; Tang, Y.; Qian, F. Opportunities and Challenges of Artificial Intelligence for Green Manufacturing in the Process Industry. Engineering 2019, 5, 995–1002. [Google Scholar] [CrossRef]
- Horwath, J.P.; Zakharov, D.N.; Mégret, R.; Stach, E.A. Understanding important features of deep learning models for segmentation of high-resolution transmission electron microscopy images. NPJ Comput. Mater. 2020, 6, 108. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P. Artificial Intelligence in Nephrology: Core Concepts, Clinical Applications, and Perspectives. Am. J. Kidney Dis. 2019, 74, 803–810. [Google Scholar] [CrossRef]
- Sumit, S. A Comprehensive Guide to Convolutional Neural Networks—The ELI5 Way. 2018. Available online: https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53 (accessed on 1 May 2022).
- Monshi, M.M.A.; Poon, J.; Chung, V. Deep learning in generating radiology reports: A survey. Artif. Intell. Med. 2020, 106, 101878. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, R.; Gupta, S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Zhang, D.Y. Artificial intelligence and computational pathology. Lab. Investig. 2021, 101, 412–422. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peikari, M.; Salama, S.; Nofech-Mozes, S.; Martel, A.L. A Cluster-then-label Semi-supervised Learning Approach for Pathology Image Classification. Sci. Rep. 2018, 8, 7193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yang, D.M.; Rong, R.; Zhan, X.; Xiao, G. Pathology Image Analysis Using Segmentation Deep Learning Algorithms. Am. J. Pathol. 2019, 189, 1686–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Cruz-Roa, A.; Basavanhally, A.; Gilmore, H.; Shih, N.; Feldman, M.; Tomaszewski, J.; Gonzalez, F.; Madabhushi, A. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J. Med. Imaging 2014, 1, 034003. [Google Scholar] [CrossRef]
- Le, E.P.V.; Wang, Y.; Huang, Y.; Hickman, S.; Gilbert, F.J. Artificial intelligence in breast imaging. Clin. Radiol. 2019, 74, 357–366. [Google Scholar] [CrossRef]
- Lee, K.M.; Street, W.N. Model-based detection, segmentation, and classification for image analysis using on-line shape learning. Mach. Vis. Appl. 2003, 13, 222–233. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Yao, K.; Singh, A.; Sridhar, K.; Blau, J.L.; Ohgami, R.S. Artificial Intelligence in Pathology: A Simple and Practical Guide. Adv. Anat. Pathol. 2020, 27, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.W.; Lei, S.T.; Zhu, M.M.; Li, R.Y.; Yue, J.Y.; Chen, J.J.; Li, Z.W.; Gong, J.M.; Lin, D.R.; Wu, X.H.; et al. Improving the Generalizability of Infantile Cataracts Detection via Deep Learning-Based Lens Partition Strategy and Multicenter Datasets. Front. Med. 2021, 8, 664023. [Google Scholar] [CrossRef] [PubMed]
- Hamidinekoo, A.; Denton, E.; Rampun, A.; Honnor, K.; Zwiggelaar, R. Deep learning in mammography and breast histology, an overview and future trends. Med. Image Anal. 2018, 47, 45–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, P.M.; Robinson, E.J.; Sarin Pradhan, J.; Gartrell-Corrado, R.D.; Rohr, B.R.; Trager, M.H.; Geskin, L.J.; Kluger, H.M.; Wong, P.F.; Acs, B.; et al. Deep Learning Based on Standard H&E Images of Primary Melanoma Tumors Identifies Patients at Risk for Visceral Recurrence and Death. Clin. Cancer Res. 2020, 26, 1126–1134. [Google Scholar] [CrossRef] [Green Version]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef] [Green Version]
- Lv, J.C.; Zhang, L.X. Prevalence and Disease Burden of Chronic Kidney Disease. Adv. Exp. Med. Biol. 2019, 1165, 3–15. [Google Scholar] [CrossRef]
- Xie, W.; Xu, J.; Xie, Y.; Lin, Z.; Xu, X.; Zhang, X.; Zhang, Y. Adequacy and complication rates of percutaneous renal biopsy with 18- vs. 16-gauge needles in native kidneys in Chinese individuals. BMC Nephrol. 2020, 21, 337. [Google Scholar] [CrossRef]
- Hermsen, M.; de Bel, T.; den Boer, M.; Steenbergen, E.J.; Kers, J.; Florquin, S.; Roelofs, J.; Stegall, M.D.; Alexander, M.P.; Smith, B.H.; et al. Deep Learning-Based Histopathologic Assessment of Kidney Tissue. J. Am. Soc. Nephrol. JASN 2019, 30, 1968–1979. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic Kidney Disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef]
- Kannan, S.; Morgan, L.A.; Liang, B.; Cheung, M.G.; Lin, C.Q.; Mun, D.; Nader, R.G.; Belghasem, M.E.; Henderson, J.M.; Francis, J.M.; et al. Segmentation of Glomeruli within Trichrome Images Using Deep Learning. Kidney Int. Rep. 2019, 4, 955–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, A.Z.; Palmer, M.; Merlino, L.; Troost, J.P.; Gasim, A.; Bagnasco, S.; Avila-Casado, C.; Johnstone, D.; Hodgin, J.B.; Conway, C.; et al. The Application of Digital Pathology to Improve Accuracy in Glomerular Enumeration in Renal Biopsies. PLoS ONE 2016, 11, e0156441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, O.; Yacoub, R.; Jain, S.; Tomaszewski, J.E.; Sarder, P. Multi-radial LBP Features as a Tool for Rapid Glomerular Detection and Assessment in Whole Slide Histopathology Images. Sci. Rep. 2018, 8, 2032. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.S. Pathology of IgA nephropathy. Nat. Rev. Nephrol. 2014, 10, 445–454. [Google Scholar] [CrossRef]
- Bajema, I.M.; Wilhelmus, S.; Alpers, C.E.; Bruijn, J.A.; Colvin, R.B.; Cook, H.T.; D’Agati, V.D.; Ferrario, F.; Haas, M.; Jennette, J.C.; et al. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: Clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int. 2018, 93, 789–796. [Google Scholar] [CrossRef]
- Chagas, P.; Souza, L.; Araújo, I.; Aldeman, N.; Duarte, A.; Angelo, M.; Dos-Santos, W.L.C.; Oliveira, L. Classification of glomerular hypercellularity using convolutional features and support vector machine. Artif. Intell. Med. 2020, 103, 101808. [Google Scholar] [CrossRef] [Green Version]
- Risdon, R.A.; Turner, D.R. Atlas of Renal Pathology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 2. [Google Scholar]
- Kolachalama, V.B.; Singh, P.; Lin, C.Q.; Mun, D.; Belghasem, M.E.; Henderson, J.M.; Francis, J.M.; Salant, D.J.; Chitalia, V.C. Association of Pathological Fibrosis with Renal Survival Using Deep Neural Networks. Kidney Int. Rep. 2018, 3, 464–475. [Google Scholar] [CrossRef] [Green Version]
- Liapis, H.; Gaut, J.P.; Klein, C.; Bagnasco, S.; Kraus, E.; Farris, A.B., 3rd; Honsova, E.; Perkowska-Ptasinska, A.; David, D.; Goldberg, J.; et al. Banff Histopathological Consensus Criteria for Preimplantation Kidney Biopsies. Am. J. Transplant. 2017, 17, 140–150. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, W.; Dong, B.; Mei, K.; Zhu, C.; Liu, J.; Cai, M.; Yan, Y.; Wang, G.; Zuo, L.; et al. A Deep Learning-Based Approach for Glomeruli Instance Segmentation from Multistained Renal Biopsy Pathologic Images. Am. J. Pathol. 2021, 191, 1431–1441. [Google Scholar] [CrossRef]
- Lutnick, B.; Ginley, B.; Jen, K.Y.; Dong, W.; Sarder, P. Generative modeling for label-free glomerular modeling and classification. Proc. SPIE Int. Soc. Opt. Eng. 2020, 11320, 1132007. [Google Scholar]
- Lu, Y.; Yang, H.; Asad, Z.; Zhu, Z.; Yao, T.; Xu, J.; Fogo, A.B.; Huo, Y. Holistic fine-grained global glomerulosclerosis characterization: From detection to unbalanced classification. J. Med. Imaging 2022, 9, 014005. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.A.; Bindzus, J.N.; Voigt, J.; Runz, M.; Hertjens, S.; Gaida, M.M.; Popovic, Z.V.; Porubsky, S. Assessment of glomerular morphological patterns by deep learning algorithms. J. Nephrol. 2022, 35, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Kawazoe, Y.; Shimamoto, K.; Shinohara, E.; Tsukamoto, T.; Shintani-Domoto, Y.; Nagasu, H.; Uozaki, H.; Ushiku, T.; Nangaku, M.; et al. Glomerular Classification Using Convolutional Neural Networks Based on Defined Annotation Criteria and Concordance Evaluation among Clinicians. Kidney Int. Rep. 2021, 6, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, M.; Wu, Y.; Hao, F.; Wang, C.; Han, W.; Niu, D.; Zheng, W. Classification of renal biopsy direct immunofluorescence image using multiple attention convolutional neural network. Comput. Methods Programs Biomed. 2022, 214, 106532. [Google Scholar] [CrossRef]
- López, J.I.; Larrinaga, G.; Kuroda, N.; Angulo, J.C. The normal and pathologic renal medulla: A comprehensive overview. Pathol.-Res. Pract. 2015, 211, 271–280. [Google Scholar] [CrossRef]
- Roufosse, C.; Simmonds, N.; Clahsen-van Groningen, M.; Haas, M.; Henriksen, K.J.; Horsfield, C.; Loupy, A.; Mengel, M.; Perkowska-Ptasińska, A.; Rabant, M.; et al. A 2018 Reference Guide to the Banff Classification of Renal Allograft Pathology. Transplantation 2018, 102, 1795–1814. [Google Scholar] [CrossRef]
- Fenstermaker, M.; Tomlins, S.A.; Singh, K.; Wiens, J.; Morgan, T.M. Development and Validation of a Deep-learning Model to Assist with Renal Cell Carcinoma Histopathologic Interpretation. Urology 2020, 144, 152–157. [Google Scholar] [CrossRef]
- Bukowy, J.D.; Dayton, A.; Cloutier, D.; Manis, A.D.; Staruschenko, A.; Lombard, J.H.; Solberg Woods, L.C.; Beard, D.A.; Cowley, A.W., Jr. Region-Based Convolutional Neural Nets for Localization of Glomeruli in Trichrome-Stained Whole Kidney Sections. J. Am. Soc. Nephrol. JASN 2018, 29, 2081–2088. [Google Scholar] [CrossRef] [Green Version]
- Sheehan, S.M.; Korstanje, R. Automatic glomerular identification and quantification of histological phenotypes using image analysis and machine learning. Am. J. Physiol. Ren. Physiol. 2018, 315, F1644–F1651. [Google Scholar] [CrossRef] [Green Version]
- Wilbur, D.C.; Smith, M.L.; Cornell, L.D.; Andryushkin, A.; Pettus, J.R. Automated identification of glomeruli and synchronised review of special stains in renal biopsies by machine learning and slide registration: A cross-institutional study. Histopathology 2021, 79, 499–508. [Google Scholar] [CrossRef]
- Wilbur, D.C.; Pettus, J.R.; Smith, M.L.; Cornell, L.D.; Andryushkin, A.; Wingard, R.; Wirch, E. Using Image Registration and Machine Learning to Develop a Workstation Tool for Rapid Analysis of Glomeruli in Medical Renal Biopsies. J. Pathol. Inform. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Barros, G.O.; Navarro, B.; Duarte, A.; Dos-Santos, W.L.C. PathoSpotter-K: A computational tool for the automatic identification of glomerular lesions in histological images of kidneys. Sci. Rep. 2017, 7, 46769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bueno, G.; Fernandez-Carrobles, M.M.; Gonzalez-Lopez, L.; Deniz, O. Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput. Methods Programs Biomed. 2020, 184, 105273. [Google Scholar] [CrossRef] [PubMed]
- Gallego, J.; Swiderska-Chadaj, Z.; Markiewicz, T.; Yamashita, M.; Gabaldon, M.A.; Gertych, A. A U-Net based framework to quantify glomerulosclerosis in digitized PAS and H&E stained human tissues. Comput. Med. Imaging Graph. 2021, 89, 101865. [Google Scholar] [CrossRef]
- Pesce, F.; Albanese, F.; Mallardi, D.; Rossini, M.; Pasculli, G.; Suavo-Bulzis, P.; Granata, A.; Brunetti, A.; Cascarano, G.D.; Bevilacqua, V.; et al. Identification of glomerulosclerosis using IBM Watson and shallow neural networks. J. Nephrol. 2022, 35, 1235–1242. [Google Scholar] [CrossRef]
- Suavo-Bulzis, P.; Albanese, F.; Mallardi, D.; Debitonto, F.S.; Lemma, R.; Granatiero, A.; Spadavecchia, M.; Cascarano, G.D.; Bevilacqua, V.; Gesualdo, L. P0119 ARTIFICIAL INTELLIGENCE IN RENAL PATHOLOGY: IBM WATSON FOR THE IDENTIFICATION OF GLOMERULOSCLEROSIS. Nephrol. Dial. Transplant. 2020, 35, gfaa142.P0119. [Google Scholar] [CrossRef]
- Cascarano, G.D.; Debitonto, F.S.; Lemma, R.; Brunetti, A.; Buongiorno, D.; De Feudis, I.; Guerriero, A.; Venere, U.; Matino, S.; Rocchetti, M.T.; et al. A neural network for glomerulus classification based on histological images of kidney biopsy. BMC Med. Inform. Decis. Mak. 2021, 21, 300. [Google Scholar] [CrossRef]
- Marsh, J.N.; Matlock, M.K.; Kudose, S.; Liu, T.C.; Stappenbeck, T.S.; Gaut, J.P.; Swamidass, S.J. Deep Learning Global Glomerulosclerosis in Transplant Kidney Frozen Sections. IEEE Trans. Med. Imaging 2018, 37, 2718–2728. [Google Scholar] [CrossRef]
- Li, X.; Davis, R.C.; Xu, Y.; Wang, Z.; Souma, N.; Sotolongo, G.; Bell, J.; Ellis, M.; Howell, D.; Shen, X.; et al. Deep learning segmentation of glomeruli on kidney donor frozen sections. J. Med. Imaging 2021, 8, 067501. [Google Scholar] [CrossRef]
- Marsh, J.N.; Liu, T.C.; Wilson, P.C.; Swamidass, S.J.; Gaut, J.P. Development and Validation of a Deep Learning Model to Quantify Glomerulosclerosis in Kidney Biopsy Specimens. JAMA Netw. Open 2021, 4, e2030939. [Google Scholar] [CrossRef]
- Uchino, E.; Suzuki, K.; Sato, N.; Kojima, R.; Tamada, Y.; Hiragi, S.; Yokoi, H.; Yugami, N.; Minamiguchi, S.; Haga, H.; et al. Classification of glomerular pathological findings using deep learning and nephrologist-AI collective intelligence approach. Int. J. Med. Inform. 2020, 141, 104231. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.K.; Lee, C.Y.; Wang, H.S.; Huang, S.C.; Liang, P.I.; Chen, J.S.; Kuo, C.F.; Tu, K.H.; Yeh, C.Y.; Chen, T.D. Glomerular Disease Classification and Lesion Identification by Machine Learning. Biomed. J. 2021; S2319-4170(21)00111-6, in press. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Li, F.; Tang, P.; Zhang, G.; Zeng, C.; Xie, G.; Liu, Z.; Yang, G. Automatic Fine-grained Glomerular Lesion Recognition in Kidney Pathology. Pattern Recognit. 2022, 127, 108648. [Google Scholar] [CrossRef]
- Sheehan, S.; Mawe, S.; Cianciolo, R.E.; Korstanje, R.; Mahoney, J.M. Detection and Classification of Novel Renal Histologic Phenotypes Using Deep Neural Networks. Am. J. Pathol. 2019, 189, 1786–1796. [Google Scholar] [CrossRef]
- Bouteldja, N.; Klinkhammer, B.M.; Bülow, R.D.; Droste, P.; Otten, S.W.; Freifrau von Stillfried, S.; Moellmann, J.; Sheehan, S.M.; Korstanje, R.; Menzel, S.; et al. Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology. J. Am. Soc. Nephrol. JASN 2021, 32, 52–68. [Google Scholar] [CrossRef]
- Jayapandian, C.P.; Chen, Y.; Janowczyk, A.R.; Palmer, M.B.; Cassol, C.A.; Sekulic, M.; Hodgin, J.B.; Zee, J.; Hewitt, S.M.; O’Toole, J.; et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 2021, 99, 86–101. [Google Scholar] [CrossRef]
- Govind, D.; Becker, J.U.; Miecznikowski, J.; Rosenberg, A.Z.; Dang, J.; Tharaux, P.L.; Yacoub, R.; Thaiss, F.; Hoyer, P.F.; Manthey, D.; et al. PodoSighter: A Cloud-Based Tool for Label-Free Podocyte Detection in Kidney Whole-Slide Images. J. Am. Soc. Nephrol. JASN 2021, 32, 2795–2813. [Google Scholar] [CrossRef]
- Marostica, E.; Barber, R.; Denize, T.; Kohane, I.S.; Signoretti, S.; Golden, J.A.; Yu, K.H. Development of a Histopathology Informatics Pipeline for Classification and Prediction of Clinical Outcomes in Subtypes of Renal Cell Carcinoma. Clin. Cancer Res. 2021, 27, 2868–2878. [Google Scholar] [CrossRef]
- Tabibu, S.; Vinod, P.K.; Jawahar, C.V. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci. Rep. 2019, 9, 10509. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.D.; Ren, B.; Richards, R.; Suriawinata, M.; Tomita, N.; Hassanpour, S. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci. Rep. 2021, 11, 7080. [Google Scholar] [CrossRef]
- Hewitson, T.D.; Holt, S.G.; Smith, E.R. Progression of Tubulointerstitial Fibrosis and the Chronic Kidney Disease Phenotype—Role of Risk Factors and Epigenetics. Front. Pharmacol. 2017, 8, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuiblet, V.; Fere, M.; Gobinet, C.; Birembaut, P.; Piot, O.; Rieu, P. Renal Graft Fibrosis and Inflammation Quantification by an Automated Fourier-Transform Infrared Imaging Technique. J. Am. Soc. Nephrol. JASN 2016, 27, 2382–2391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stribos, E.G.D.; Nielsen, S.H.; Brix, S.; Karsdal, M.A.; Seelen, M.A.; van Goor, H.; Bakker, S.J.L.; Olinga, P.; Mutsaers, H.A.M.; Genovese, F. Non-invasive quantification of collagen turnover in renal transplant recipients. PLoS ONE 2017, 12, e0175898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.S.; Li, Y.P.; Ni, H.F. Morphology and Evaluation of Renal Fibrosis. Adv. Exp. Med. Biol. 2019, 1165, 17–36. [Google Scholar] [CrossRef]
- Furness, P.N.; Taub, N. International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project. Kidney Int. 2001, 60, 1998–2012. [Google Scholar] [CrossRef] [Green Version]
- Huo, Y.; Deng, R.; Liu, Q.; Fogo, A.B.; Yang, H. AI applications in renal pathology. Kidney Int. 2021, 99, 1309–1320. [Google Scholar] [CrossRef]
- Ginley, B.; Jen, K.Y.; Han, S.S.; Rodrigues, L.; Jain, S.; Fogo, A.B.; Zuckerman, J.; Walavalkar, V.; Miecznikowski, J.C.; Wen, Y.; et al. Automated Computational Detection of Interstitial Fibrosis, Tubular Atrophy, and Glomerulosclerosis. J. Am. Soc. Nephrol. JASN 2021, 32, 837–850. [Google Scholar] [CrossRef]
- Marechal, E.; Jaugey, A.; Tarris, G.; Paindavoine, M.; Seibel, J.; Martin, L.; Funes de la Vega, M.; Crepin, T.; Ducloux, D.; Zanetta, G.; et al. Automatic Evaluation of Histological Prognostic Factors Using Two Consecutive Convolutional Neural Networks on Kidney Samples. Clin. J. Am. Soc. Nephrol. 2022, 17, 260–270. [Google Scholar] [CrossRef]
- Yi, Z.; Salem, F.; Menon, M.C.; Keung, K.; Xi, C.; Hultin, S.; Haroon Al Rasheed, M.R.; Li, L.; Su, F.; Sun, Z.; et al. Deep learning identified pathological abnormalities predictive of graft loss in kidney transplant biopsies. Kidney Int. 2022, 101, 288–298. [Google Scholar] [CrossRef]
- Shi, H.; Jia, J.; Li, D.; Wei, L.; Shang, W.; Zheng, Z. Blood oxygen level dependent magnetic resonance imaging for detecting pathological patterns in lupus nephritis patients: A preliminary study using a decision tree model. BMC Nephrol. 2018, 19, 33. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.Y.; Huang, D.L.; Dang, X.Q.; Yi, Z.W. Lupus glomerulonephritis in 788 Chinese children: A multi-centre clinical and histopathological analysis based on 549 renal biopsies. Paediatr. Int. Child Health 2017, 37, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Faurschou, M.; Starklint, H.; Halberg, P.; Jacobsen, S. Prognostic factors in lupus nephritis: Diagnostic and therapeutic delay increases the risk of terminal renal failure. J. Rheumatol. 2006, 33, 1563–1569. [Google Scholar] [PubMed]
- Dasari, S.; Chakraborty, A.; Truong, L.; Mohan, C. A Systematic Review of Interpathologist Agreement in Histologic Classification of Lupus Nephritis. Kidney Int. Rep. 2019, 4, 1420–1425. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Zhang, X.; Ding, J.; Zhang, D.; Cui, J.; Fu, X.; Han, J.; Zhu, P. Deep Learning-Based Artificial Intelligence System for Automatic Assessment of Glomerular Pathological Findings in Lupus Nephritis. Diagnostics 2021, 11, 1983. [Google Scholar] [CrossRef]
- Kudose, S.; Santoriello, D.; Bomback, A.S.; Stokes, M.B.; D’Agati, V.D.; Markowitz, G.S. Sensitivity and Specificity of Pathologic Findings to Diagnose Lupus Nephritis. Clin. J. Am. Soc. Nephrol. 2019, 14, 1605–1615. [Google Scholar] [CrossRef]
- Pan, S.; Fu, Y.; Chen, P.; Liu, J.; Liu, W.; Wang, X.; Cai, G.; Yin, Z.; Wu, J.; Tang, L.; et al. Multi-Task Learning-Based Immunofluorescence Classification of Kidney Disease. Int. J. Environ. Res. Public Health 2021, 18, 10798. [Google Scholar] [CrossRef]
- Helget, L.N.; Dillon, D.J.; Wolf, B.; Parks, L.P.; Self, S.E.; Bruner, E.T.; Oates, E.E.; Oates, J.C. Development of a lupus nephritis suboptimal response prediction tool using renal histopathological and clinical laboratory variables at the time of diagnosis. Lupus Sci. Med. 2021, 8, e000489. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, S.; Chen, T.; Liang, D.; Yang, J.; Zeng, C.; Li, X.; Xie, G.; Liu, Z. Machine Learning for Prediction and Risk Stratification of Lupus Nephritis Renal Flare. Am. J. Nephrol. 2021, 52, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.X.; Regier, E.E.; Close, K.L. International Diabetes Federation World Diabetes Congress 2015. J. Diabetes 2016, 8, 300–302. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.; Mao, X.; Zhang, Z.; Wu, H. Classification and Differential Diagnosis of Diabetic Nephropathy. J. Diabetes Res. 2017, 2017, 8637138. [Google Scholar] [CrossRef] [Green Version]
- Tervaert, T.W.; Mooyaart, A.L.; Amann, K.; Cohen, A.H.; Cook, H.T.; Drachenberg, C.B.; Ferrario, F.; Fogo, A.B.; Haas, M.; de Heer, E.; et al. Pathologic classification of diabetic nephropathy. J. Am. Soc. Nephrol. JASN 2010, 21, 556–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginley, B.; Lutnick, B.; Jen, K.Y.; Fogo, A.B.; Jain, S.; Rosenberg, A.; Walavalkar, V.; Wilding, G.; Tomaszewski, J.E.; Yacoub, R.; et al. Computational Segmentation and Classification of Diabetic Glomerulosclerosis. J. Am. Soc. Nephrol. 2019, 30, 1953–1967. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Takahashi, K.; Sang, Y.; Fukushima, K.; Tsuji, K.; Wada, J. Deep Learning Could Diagnose Diabetic Nephropathy with Renal Pathological Immunofluorescent Images. Diagnostics 2020, 10, 466. [Google Scholar] [CrossRef]
- Hacking, S.; Bijol, V. Deep learning for the classification of medical kidney disease: A pilot study for electron microscopy. Ultrastruct. Pathol. 2021, 45, 118–127. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, G. Natural history of idiopathic IgA nephropathy and factors predictive of disease outcome. Semin. Nephrol. 2004, 24, 179–196. [Google Scholar] [CrossRef]
- Bülow, R.D.; Dimitrov, D.; Boor, P.; Saez-Rodriguez, J. How will artificial intelligence and bioinformatics change our understanding of IgA Nephropathy in the next decade? Semin. Immunopathol. 2021, 43, 739–752. [Google Scholar] [CrossRef]
- Wyatt, R.J.; Julian, B.A. IgA nephropathy. N. Engl. J. Med. 2013, 368, 2402–2414. [Google Scholar] [CrossRef] [Green Version]
- Trimarchi, H.; Barratt, J.; Cattran, D.C.; Cook, H.T.; Coppo, R.; Haas, M.; Liu, Z.H.; Roberts, I.S.; Yuzawa, Y.; Zhang, H.; et al. Oxford Classification of IgA nephropathy 2016: An update from the IgA Nephropathy Classification Working Group. Kidney Int. 2017, 91, 1014–1021. [Google Scholar] [CrossRef] [Green Version]
- Coppo, R.; Troyanov, S.; Bellur, S.; Cattran, D.; Cook, H.T.; Feehally, J.; Roberts, I.S.; Morando, L.; Camilla, R.; Tesar, V.; et al. Validation of the Oxford classification of IgA nephropathy in cohorts with different presentations and treatments. Kidney Int. 2014, 86, 828–836. [Google Scholar] [CrossRef] [Green Version]
- Coppo, R.; D’Arrigo, G.; Tripepi, G.; Russo, M.L.; Roberts, I.S.D.; Bellur, S.; Cattran, D.; Cook, T.H.; Feehally, J.; Tesar, V.; et al. Is there long-term value of pathology scoring in immunoglobulin A nephropathy? A validation study of the Oxford Classification for IgA Nephropathy (VALIGA) update. Nephrol. Dial. Transplant. 2020, 35, 1002–1009. [Google Scholar] [CrossRef]
- Palamuthusingam, D.; Castledine, C.; Lawman, S. Outcomes of immunosuppression in IgA nephropathy based on the oxford classification. Saudi J. Kidney Dis. Transplant. 2018, 29, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Nan, Y.; Xu, F.; Lei, Q.; Li, F.; Chen, T.; Liang, S.; Hou, X.; Lv, B.; Liang, D.; et al. Identification of glomerular lesions and intrinsic glomerular cell types in kidney diseases via deep learning. J. Pathol. 2020, 252, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Uchino, E.; Kojima, R.; Sakuragi, M.; Hiragi, S.; Minamiguchi, S.; Haga, H.; Yokoi, H.; Yanagita, M.; Okuno, Y. Evaluation of Kidney Histological Images Using Unsupervised Deep Learning. Kidney Int. Rep. 2021, 6, 2445–2454. [Google Scholar] [CrossRef] [PubMed]
- Gowrishankar, S.; Gupta, Y.; Vankalakunti, M.; Gowda, K.K.; Kurien, A.A.; Jansi Prema, K.S.; Seethalekshmy, N.V.; Yesodharan, J. Correlation of Oxford MEST-C Scores with Clinical Variables for IgA Nephropathy in South India. Kidney Int. Rep. 2019, 4, 1485–1490. [Google Scholar] [CrossRef] [Green Version]
- Farris, A.B.; Vizcarra, J.; Amgad, M.; Cooper, L.A.D.; Gutman, D.; Hogan, J. Image Analysis Pipeline for Renal Allograft Evaluation and Fibrosis Quantification. Kidney Int. Rep. 2021, 6, 1878–1887. [Google Scholar] [CrossRef]
- Cicalese, P.A.; Mobiny, A.; Shahmoradi, Z.; Yi, X.F.; Mohan, C.; Nguyen, H.V. Kidney Level Lupus Nephritis Classification Using Uncertainty Guided Bayesian Convolutional Neural Networks. IEEE J. Biomed. Health 2021, 25, 315–324. [Google Scholar] [CrossRef]
- Ravi, M.; Hegadi, R.S. A new hybrid gkm segmentation for detecting pathological microscopic glomerulosclerosis image of diabetic nephropathy. J. Int. Pharm. Res. 2019, 46, 633–638. [Google Scholar]
- Purwar, S.; Tripathi, R.; Barwad, A.W.; Dinda, A.K. Detection of Mesangial hypercellularity of MEST-C score in immunoglobulin A-nephropathy using deep convolutional neural network. Multimed. Tools Appl. 2020, 79, 27683–27703. [Google Scholar] [CrossRef]
- Lee, J.; Warner, E.; Shaikhouni, S.; Bitzer, M.; Kretzler, M.; Gipson, D.; Pennathur, S.; Bellovich, K.; Bhat, Z.; Gadegbeku, C.; et al. Unsupervised machine learning for identifying important visual features through bag-of-words using histopathology data from chronic kidney disease. Sci. Rep. 2022, 12, 4832. [Google Scholar] [CrossRef]
- Ledbetter, D.; Ho, L.; Lemley, K.V. Prediction of kidney function from biopsy images using convolutional neural networks. arXiv 2017, arXiv:1702.01816. [Google Scholar]
- Alcorn, M.A.; Li, Q.; Gong, Z.; Wang, C.; Mai, L.; Ku, W.S.; Nguyen, A. Strike (with) a Pose: Neural Networks Are Easily Fooled by Strange Poses of Familiar Objects. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4840–4849. [Google Scholar]
- Nguyen, A.; Yosinski, J.; Clune, J. Deep neural networks are easily fooled: High confidence predictions for unrecognizable images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 427–436. [Google Scholar]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Hartman, D.J.; Pantanowitz, L.; McHugh, J.S.; Piccoli, A.L.; Oleary, M.J.; Lauro, G.R. Enterprise Implementation of Digital Pathology: Feasibility, Challenges, and Opportunities. J. Digit. Imaging 2017, 30, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Cassol, C.; Sharma, S. Nephrology Lagging Behind in Machine Learning Utilization. Kidney Med. 2021, 3, 693–695. [Google Scholar] [CrossRef]
- Nadkarni, G.N.; Chaudhary, K.; Coca, S.G. Machine Learning in Glomerular Diseases: Promise for Precision Medicine. Am. J. Kidney Dis. 2019, 74, 290–292. [Google Scholar] [CrossRef] [PubMed]
- Vasiljević, J.; Feuerhake, F.; Wemmert, C.; Lampert, T. Towards histopathological stain invariance by Unsupervised Domain Augmentation using generative adversarial networks. Neurocomputing 2021, 460, 277–291. [Google Scholar] [CrossRef]
- Summaira, J.; Li, X.; Shoib, A.M.; Li, S.; Abdul, J. Recent Advances and Trends in Multimodal Deep Learning: A Review. arXiv 2021, arXiv:2105.11087. [Google Scholar]
- Race, A.M.; Sutton, D.; Hamm, G.; Maglennon, G.; Morton, J.P.; Strittmatter, N.; Campbell, A.; Sansom, O.J.; Wang, Y.; Barry, S.T.; et al. Deep Learning-Based Annotation Transfer between Molecular Imaging Modalities: An Automated Workflow for Multimodal Data Integration. Anal. Chem. 2021, 93, 3061–3071. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Argyriou, A.; Evgeniou, T.; Pontil, M. Multi-task feature learning. Adv. Neural Inf. Processing Syst. 2006, 19, 41–48. [Google Scholar]
- Jin, C.; Yu, H.; Ke, J.; Ding, P.; Yi, Y.; Jiang, X.; Duan, X.; Tang, J.; Chang, D.T.; Wu, X.; et al. Predicting treatment response from longitudinal images using multi-task deep learning. Nat. Commun. 2021, 12, 1851. [Google Scholar] [CrossRef]
- Aeffner, F.; Zarella, M.D.; Buchbinder, N.; Bui, M.M.; Goodman, M.R.; Hartman, D.J.; Lujan, G.M.; Molani, M.A.; Parwani, A.V.; Lillard, K.; et al. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J. Pathol. Inform. 2019, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Chen, T.; Li, Y.; Chen, T.; Li, X.; Liu, Z. Artificial Intelligence in Nephrology: How Can Artificial Intelligence Augment Nephrologists’ Intelligence? Kidney Dis. 2020, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wen, Q.; Jin, L.; Chen, W. Artificial Intelligence-Assisted Renal Pathology: Advances and Prospects. J. Clin. Med. 2022, 11, 4918. https://doi.org/10.3390/jcm11164918
Wang Y, Wen Q, Jin L, Chen W. Artificial Intelligence-Assisted Renal Pathology: Advances and Prospects. Journal of Clinical Medicine. 2022; 11(16):4918. https://doi.org/10.3390/jcm11164918
Chicago/Turabian StyleWang, Yiqin, Qiong Wen, Luhua Jin, and Wei Chen. 2022. "Artificial Intelligence-Assisted Renal Pathology: Advances and Prospects" Journal of Clinical Medicine 11, no. 16: 4918. https://doi.org/10.3390/jcm11164918
APA StyleWang, Y., Wen, Q., Jin, L., & Chen, W. (2022). Artificial Intelligence-Assisted Renal Pathology: Advances and Prospects. Journal of Clinical Medicine, 11(16), 4918. https://doi.org/10.3390/jcm11164918







