MobileSkin: Classification of Skin Lesion Images Acquired Using Mobile Phone-Attached Hand-Held Dermoscopes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mobile Dermoscopy Dataset
2.1.1. Data Collection
2.1.2. Data Augmentation
2.2. Deep Learning Model
2.2.1. Deep Learning Architectures
2.2.2. Transfer Learning and Fine-Tuning
2.2.3. Network Implementation
2.2.4. Testing
3. Results
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Cancer Facts and Figures 2021. 2021. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2021/cancer-facts-and-figures-2021.pdf (accessed on 15 July 2022).
- Oliveira, R.B.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R. Computational methods for pigmented skin lesion classification in images: Review and future trends. Neural Comput. Appl. 2018, 29, 613–636. [Google Scholar] [CrossRef]
- Jerant, A.F.; Johnson, J.T.; Sheridan, C.D.; Caffrey, T.J. Early detection and treatment of skin cancer. Am. Fam. Physician 2000, 62, 357–368. [Google Scholar]
- Rigel, D.S.; Friedman, R.J.; Kopf, A.W. The incidence of malignant melanoma in the United States: Issues as we approach the 21st century. J. Am. Acad. Dermatol. 1996, 34, 839–847. [Google Scholar] [CrossRef]
- Binder, M.; Schwarz, M.; Winkler, A.; Steiner, A.; Kaider, A.; Wolff, K.; Pehamberger, H. Epiluminescence microscopy: A useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Arch. Dermatol. 1995, 131, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Carli, P.; Quercioli, E.; Sestini, S.; Stante, M.; Ricci, L.; Brunasso, G.; De Giorgi, V. Pattern analysis, not simplified algorithms, is the most reliable method for teaching dermoscopy for melanoma diagnosis to residents in dermatology. Br. J. Dermatol. 2003, 148, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Nachbar, F.; Stolz, W.; Merkle, T.; Cognetta, A.B.; Vogt, T.; Landthaler, M.; Bilek, P.; Braun-Falco, O.; Plewig, G. The ABCD rule of dermatoscopy: High prospective value in the diagnosis of doubtful melanocytic skin lesions. J. Am. Acad. Dermatol. 1994, 30, 551–559. [Google Scholar] [CrossRef]
- Argenziano, G.; Fabbrocini, G.; Carli, P.; De Giorgi, V.; Sammarco, E.; Delfino, M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: Comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch. Dermatol. 1998, 134, 1563–1570. [Google Scholar] [CrossRef]
- Kasmi, R.; Mokrani, K. Classification of malignant melanoma and benign skin lesions: Implementation of automatic ABCD rule. IET Image Process. 2016, 10, 448–455. [Google Scholar] [CrossRef]
- Kaluri, R.; Reddy, P. Sign gesture recognition using modified region growing algorithm and adaptive genetic fuzzy classifier. Int. J. Intell. Eng. Syst. 2016, 9, 225–233. [Google Scholar] [CrossRef]
- Kaluri, R.; Pradeep Reddy, C. A framework for sign gesture recognition using improved genetic algorithm and adaptive filter. Cogent Eng. 2016, 3, 1251730. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Damaševičius, R.; Eltoukhy, M.M. Machine learning and deep learning methods for skin lesion classification and diagnosis: A systematic review. Diagnostics 2021, 11, 1390. [Google Scholar] [CrossRef]
- Shahabi, F.; Rouhi, A.; Rastegari, R. The Performance of Deep and Conventional Machine Learning Techniques for Skin Lesion Classification. In Proceedings of the 2021 IEEE 18th International Conference on Smart Communities: Improving Quality of Life Using ICT, IoT and AI (HONET), Karachi, Pakistan, 11–13 October 2021; pp. 50–55. [Google Scholar]
- Yilmaz, A.; Demircali, A.A.; Kocaman, S.; Uvet, H. Comparison of Deep Learning and Traditional Machine Learning Techniques for Classification of Pap Smear Images. arXiv 2020, arXiv:2009.06366. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2016, 36, 994–1004. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Combalia, M.; Codella, N.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic lesions in the wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Yap, J.; Yolland, W.; Tschandl, P. Multimodal skin lesion classification using deep learning. Exp. Dermatol. 2018, 27, 1261–1267. [Google Scholar] [CrossRef]
- Jain, A.; Way, D.; Gupta, V.; Gao, Y.; de Oliveira Marinho, G.; Hartford, J.; Sayres, R.; Kanada, K.; Eng, C.; Nagpal, K.; et al. Development and Assessment of an Artificial Intelligence–Based Tool for Skin Condition Diagnosis by Primary Care Physicians and Nurse Practitioners in Teledermatology Practices. JAMA Netw. Open 2021, 4, e217249. [Google Scholar] [CrossRef]
- Yilmaz, A.; Göktay, F.; Varol, R.; Gencoglan, G.; Uvet, H. Deep convolutional neural networks for onychomycosis detection using microscopic images with KOH examination. Mycoses, 2022; Early View. [Google Scholar]
- Wadhawan, T.; Situ, N.; Rui, H.; Lancaster, K.; Yuan, X.; Zouridakis, G. Implementation of the 7-point checklist for melanoma detection on smart handheld devices. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 3180–3183. [Google Scholar]
- Börve, A.; Terstappen, K.; Sandberg, C.; Paoli, J. Mobile teledermoscopy—There’s an app for that! Dermatol. Pract. Concept. 2013, 3, 41. [Google Scholar] [PubMed]
- Hoang, L.; Lee, S.H.; Lee, E.J.; Kwon, K.R. Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. Appl. Sci. 2022, 12, 2677. [Google Scholar]
- Yilmaz, A.; Kalebasi, M.; Samoylenko, Y.; Guvenilir, M.E.; Uvet, H. Benchmarking of Lightweight Deep Learning Architectures for Skin Cancer Classification using ISIC 2017 Dataset. arXiv 2021, arXiv:2110.12270. [Google Scholar]
- Wei, L.; Ding, K.; Hu, H. Automatic skin cancer detection in dermoscopy images based on ensemble lightweight deep learning network. IEEE Access 2020, 8, 99633–99647. [Google Scholar]
- Chaturvedi, S.S.; Gupta, K.; Prasad, P.S. Skin lesion analyser: An efficient seven-way multi-class skin cancer classification using MobileNet. In Advances in Intelligent Systems and Computing, Proceedings of the International Conference on Advanced Machine Learning Technologies and Applications; Springer: Singapore, 2020; pp. 165–176. [Google Scholar]
- Torrey, L.; Shavlik, J. Transfer learning. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2010; pp. 242–264. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar]
- LeCun, Y.A.; Bottou, L.; Orr, G.B.; Müller, K.R. Efficient backprop. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012; pp. 9–48. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8697–8710. [Google Scholar]
- Sifre, L.; Mallat, S. Rigid-motion scattering for texture classification. arXiv 2014, arXiv:1403.1687. [Google Scholar]
- Zoph, B.; Le, Q.V. Neural architecture search with reinforcement learning. arXiv 2016, arXiv:1611.01578. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? arXiv 2014, arXiv:1411.1792. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Marchetti, M.A.; Codella, N.C.; Dusza, S.W.; Gutman, D.A.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, M.E.; DeFazio, J.L.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018, 78, 270–277. [Google Scholar] [CrossRef]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Fujisawa, Y.; Otomo, Y.; Ogata, Y.; Nakamura, Y.; Fujita, R.; Ishitsuka, Y.; Watanabe, R.; Okiyama, N.; Ohara, K.; Fujimoto, M. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br. J. Dermatol. 2019, 180, 373–381. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Fröhling, S.; et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur. J. Cancer 2019, 111, 148–154. [Google Scholar] [CrossRef]
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef]
- Pacheco, A.G.; Krohling, R.A. The impact of patient clinical information on automated skin cancer detection. Comput. Biol. Med. 2020, 116, 103545. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.Y.; Kreusch, J.; Lallas, A.; et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. 2019, 155, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jain, A.; Eng, C.; Way, D.H.; Lee, K.; Bui, P.; Kanada, K.; de Oliveira Marinho, G.; Gallegos, J.; Gabriele, S.; et al. A deep learning system for differential diagnosis of skin diseases. Nat. Med. 2020, 26, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Roder, L. Netron, Visualizer for neural network, deep learning, and machine learning models. 2020. Available online: https://doi.org/10.5281/zenodo.5854962 (accessed on 5 June 2022).

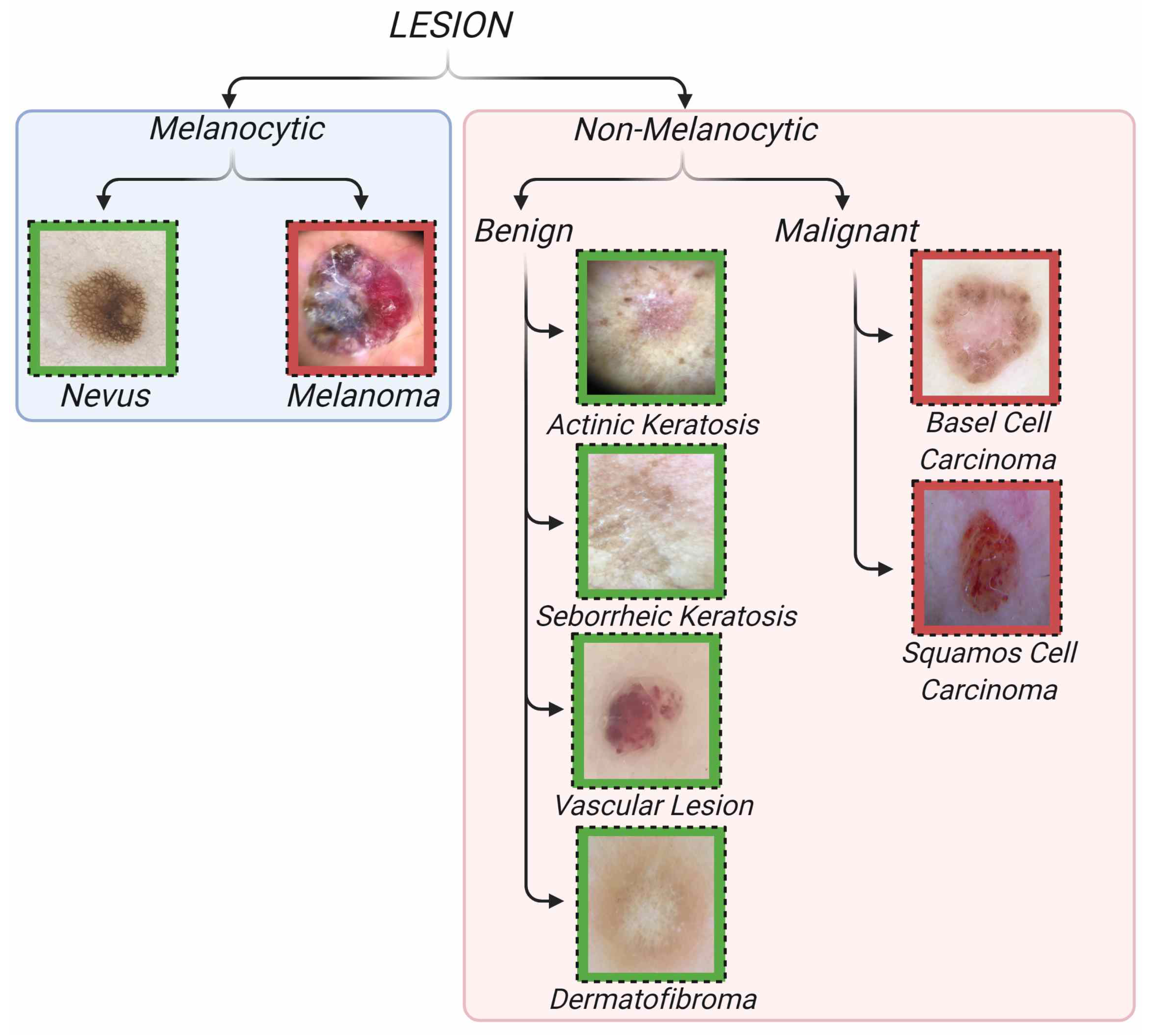




| Type | Lesion Name | Class Number | Training-Testing-Total Class Size |
|---|---|---|---|
| Non-Melanocytic Benign | Actinic Keratosis (ak) | 1 | 38-10-48 |
| Non-Melanocytic Benign | Vascular Lesion (vasc) | 2 | 160-40-200 |
| Non-Melanocytic Benign | Seborrheic Keratosis (sk) | 3 | 143-36-179 |
| Non-Melanocytic Benign | Dermatofibroma (df) | 4 | 29-7-36 |
| Non-Melanocytic Malignant | Basel Cell Carcinoma (bcc) | 5 | 188-47-235 |
| Non-Melanocytic Malignant | Squamous Cell Carcinoma (scc) | 6 | 141-35-176 |
| Melanocytic Malignant | Melanoma (mel) | 7 | 124-31-155 |
| Melanocytic Benign | Nevus (nv) | 8 | 492-123-615 |
| Total | - | - | 1315-329-1644 |
| Settings | Values |
|---|---|
| Rotation Range | 45 |
| Zoom Range | 0.2 |
| Width Shift Range | 0.2 |
| Height Shift Range | 0.2 |
| Horizontal Flip | True |
| Vertical Flip | True |
| Metric | Formula |
|---|---|
| Accuracy | |
| Precision | |
| Score |
| Metric | MobileNetV1 | MobileNetV2 | NASNetMobile | Xception |
|---|---|---|---|---|
| Accuracy | 76.96% | 89.18% | 77.21% | 89.64% |
| Precision | 77.94 | 88.13% | 78.04% | 89.99% |
| Score | 77.45% | 87.38% | 77.62% | 89.81% |
| Lesion | MobileNetV1 | MobileNetV2 | NASNetMobile | Xception |
|---|---|---|---|---|
| ak | 68.00% () | 80.00% () | 72.00% () | 66.00% () |
| vasc | 80.50% () | 90.50% () | 78.50% () | 91.00% () |
| sk | 52.78% () | 67.78% () | 56.11% () | 72.78% () |
| df | 37.14% () | 68.57% () | 40.00% () | 71.43% () |
| bcc | 65.11% () | 73.62% () | 61.70% () | 73.19% () |
| scc | 65.14% () | 89.71% () | 65.14% () | 85.71% () |
| mel | 85.81% () | 89.03% () | 85.81% () | 87.74% () |
| nv | 91.38% () | 91.87% () | 92.52% () | 91.00% () |
| Dataset | Study | Type | Comparison with Dermatologists | Dataset Size | Class Size | Dermatologists Number |
|---|---|---|---|---|---|---|
| Hybrid 1 * | [16] | Clinic | Yes | 129,450 | 9 | 2 |
| Hybrid 2 ** | [42] | Clinic | Yes | 19,398 | 12 | 16 |
| [43] | Dermoscopic | No | 200 | 3 | - | |
| ISIC 2016 | [44,45] | Dermoscopic | Yes | 1279 | 3 | 8 |
| ISIC 2017 | [46] | Dermoscopic | No | 2750 | 3 | - |
| ISIC 2018 | [19,47,48] | Dermoscopic | Yes | 10,015 | 7 | 511 |
| ISIC 2019 | [19,20] | Dermoscopic | No | 25,331 | 8 | - |
| ISIC 2020 | [17] | Dermoscopic | No | 33,126 | 2 | - |
| Mobile Dermoscopy | Own | Dermoscopic | No | 1644 | 8 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yilmaz, A.; Gencoglan, G.; Varol, R.; Demircali, A.A.; Keshavarz, M.; Uvet, H. MobileSkin: Classification of Skin Lesion Images Acquired Using Mobile Phone-Attached Hand-Held Dermoscopes. J. Clin. Med. 2022, 11, 5102. https://doi.org/10.3390/jcm11175102
Yilmaz A, Gencoglan G, Varol R, Demircali AA, Keshavarz M, Uvet H. MobileSkin: Classification of Skin Lesion Images Acquired Using Mobile Phone-Attached Hand-Held Dermoscopes. Journal of Clinical Medicine. 2022; 11(17):5102. https://doi.org/10.3390/jcm11175102
Chicago/Turabian StyleYilmaz, Abdurrahim, Gulsum Gencoglan, Rahmetullah Varol, Ali Anil Demircali, Meysam Keshavarz, and Huseyin Uvet. 2022. "MobileSkin: Classification of Skin Lesion Images Acquired Using Mobile Phone-Attached Hand-Held Dermoscopes" Journal of Clinical Medicine 11, no. 17: 5102. https://doi.org/10.3390/jcm11175102
APA StyleYilmaz, A., Gencoglan, G., Varol, R., Demircali, A. A., Keshavarz, M., & Uvet, H. (2022). MobileSkin: Classification of Skin Lesion Images Acquired Using Mobile Phone-Attached Hand-Held Dermoscopes. Journal of Clinical Medicine, 11(17), 5102. https://doi.org/10.3390/jcm11175102






