BSG (CD147) Serum Level and Genetic Variants Are Associated with Overall Survival in Acute Myeloid Leukaemia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Cell Lines
2.2. Patients and Controls
2.3. RNA Extraction and Gene Expression Analysis
2.4. ELISA Analysis
2.5. Western Blot Analysis
2.6. DNA Isolation, SNP Selection and Genotyping
2.7. Statistical Analysis
3. Results
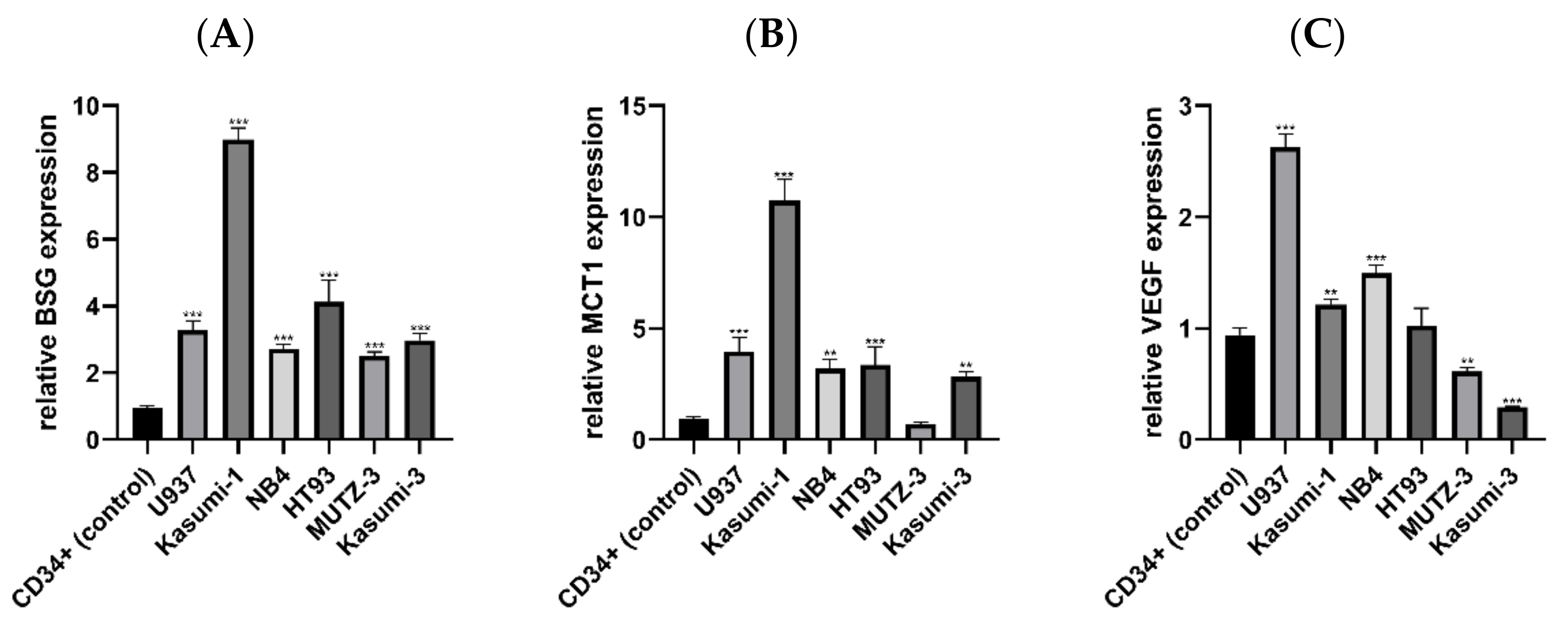
3.1. Expression of BSG, MCT1 and VEGF mRNA in AML and Control Cell Lines
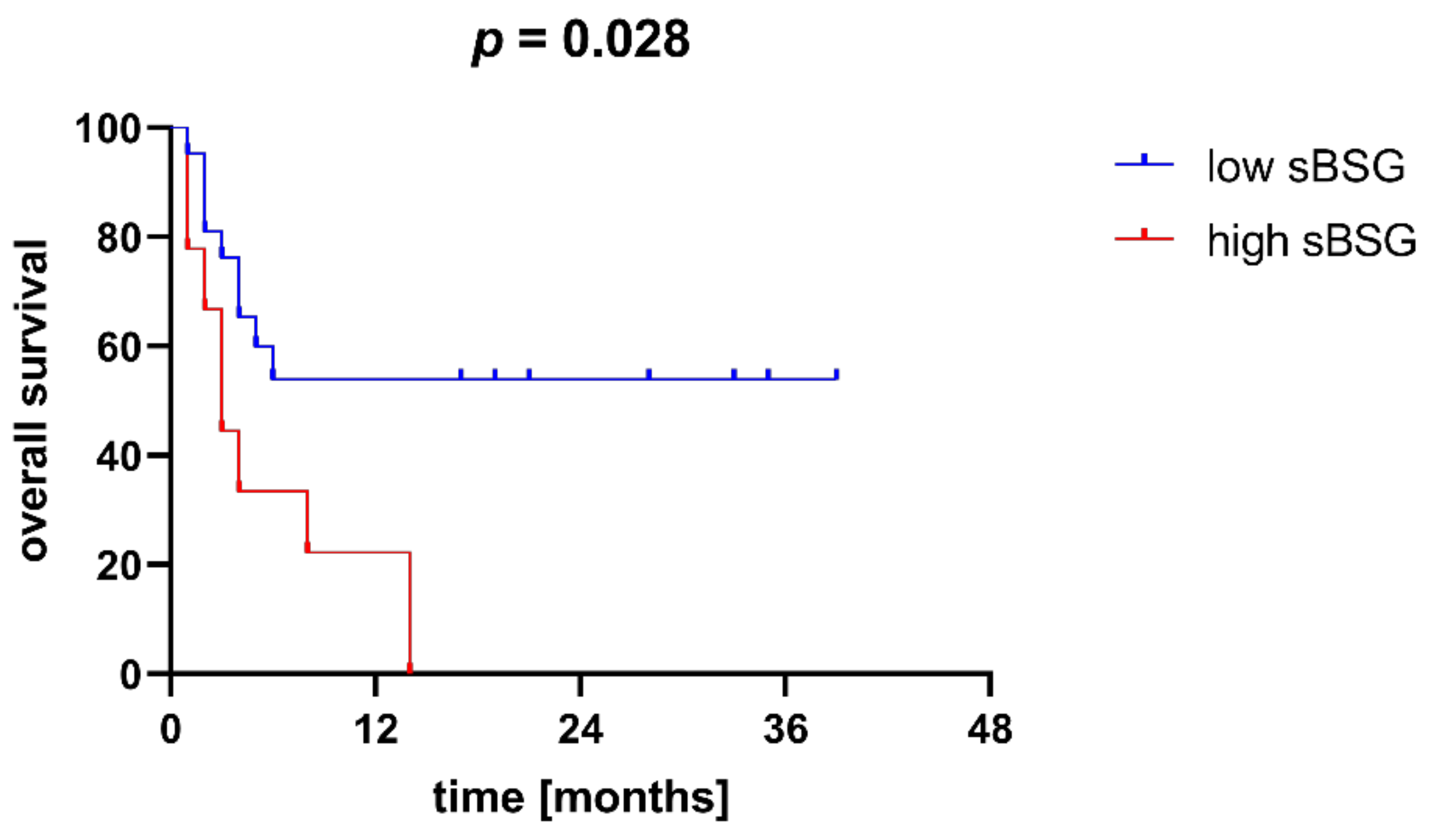
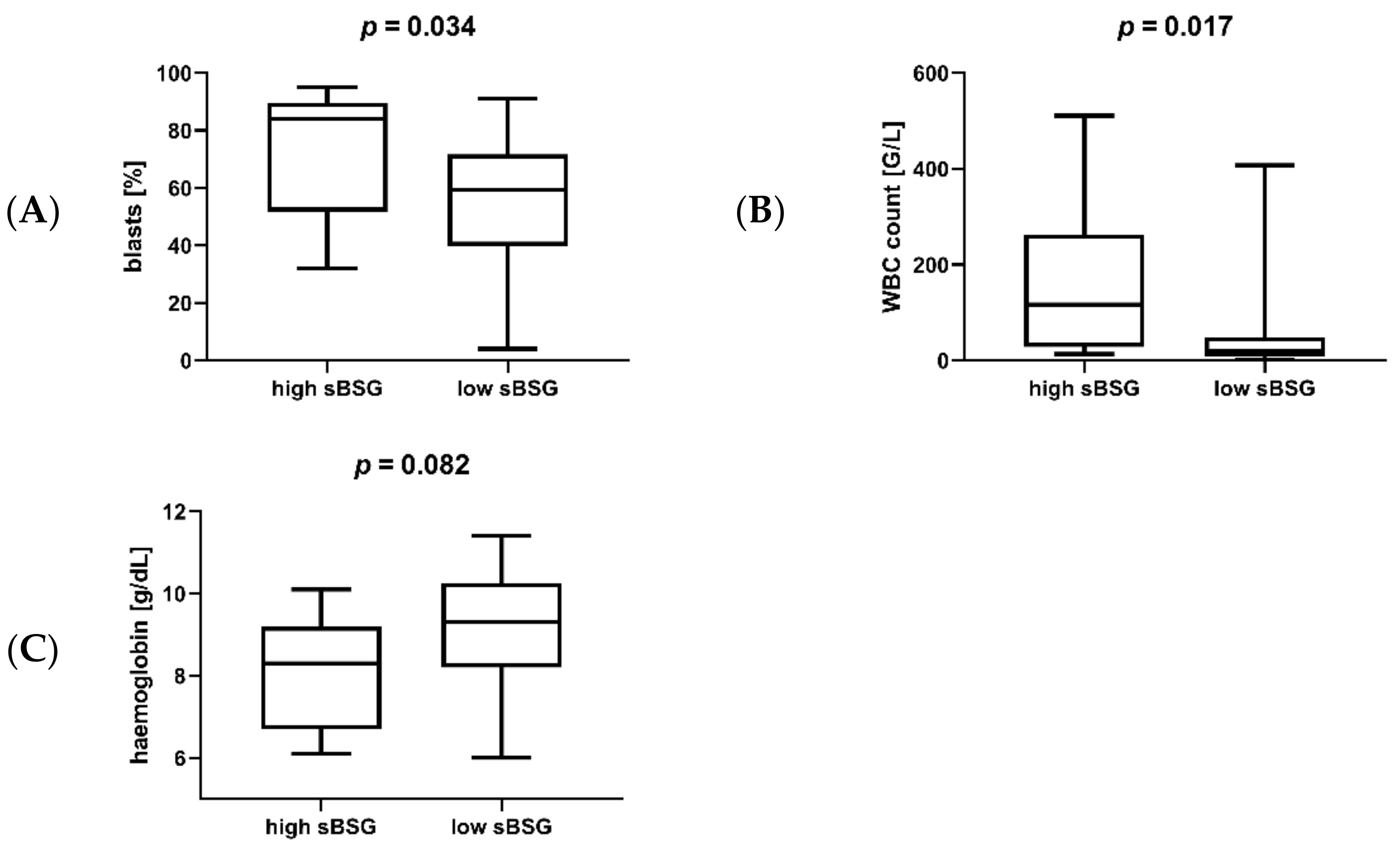
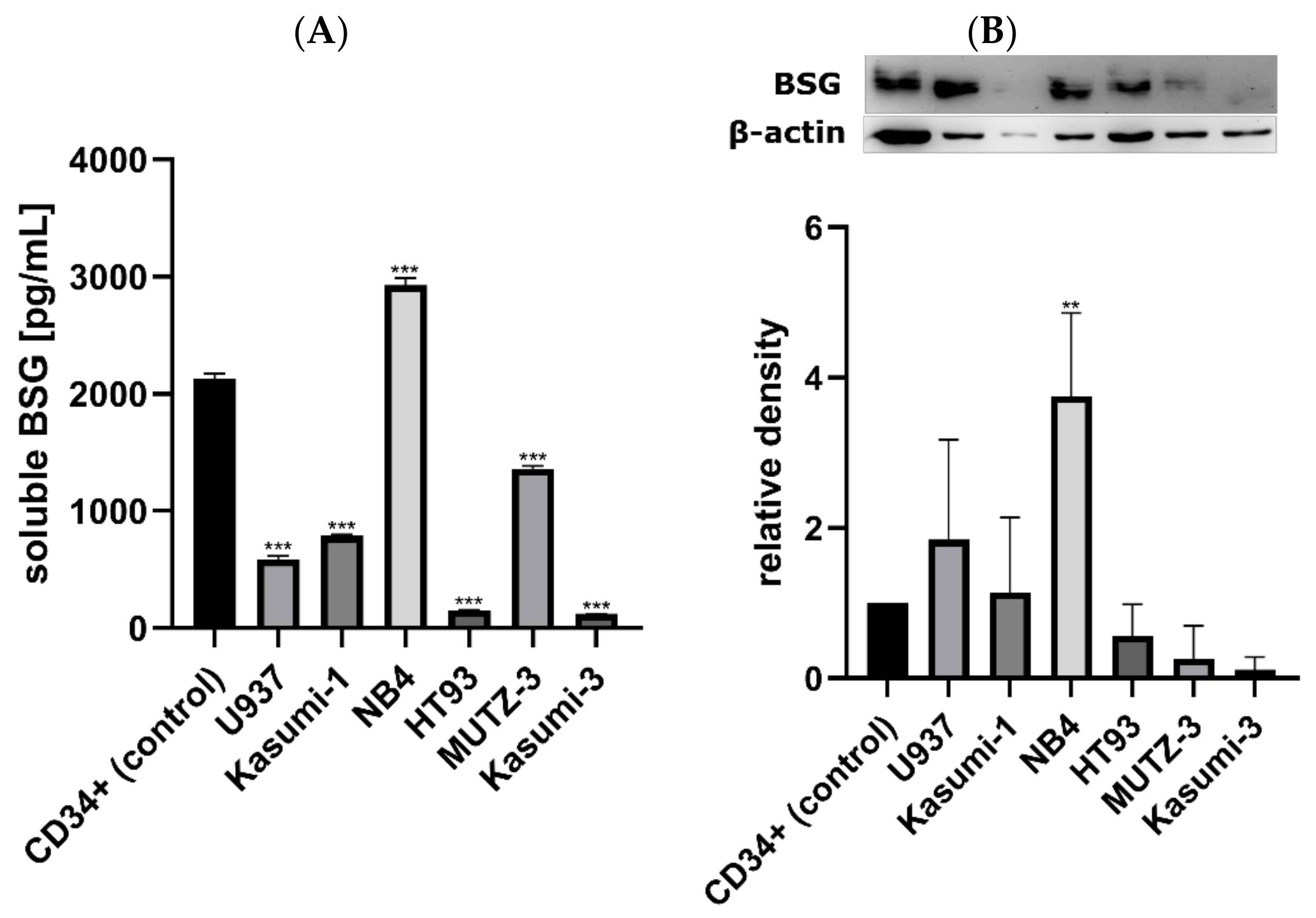
3.2. Soluble BSG in Serum as a Marker of AML Susceptibility and Survival
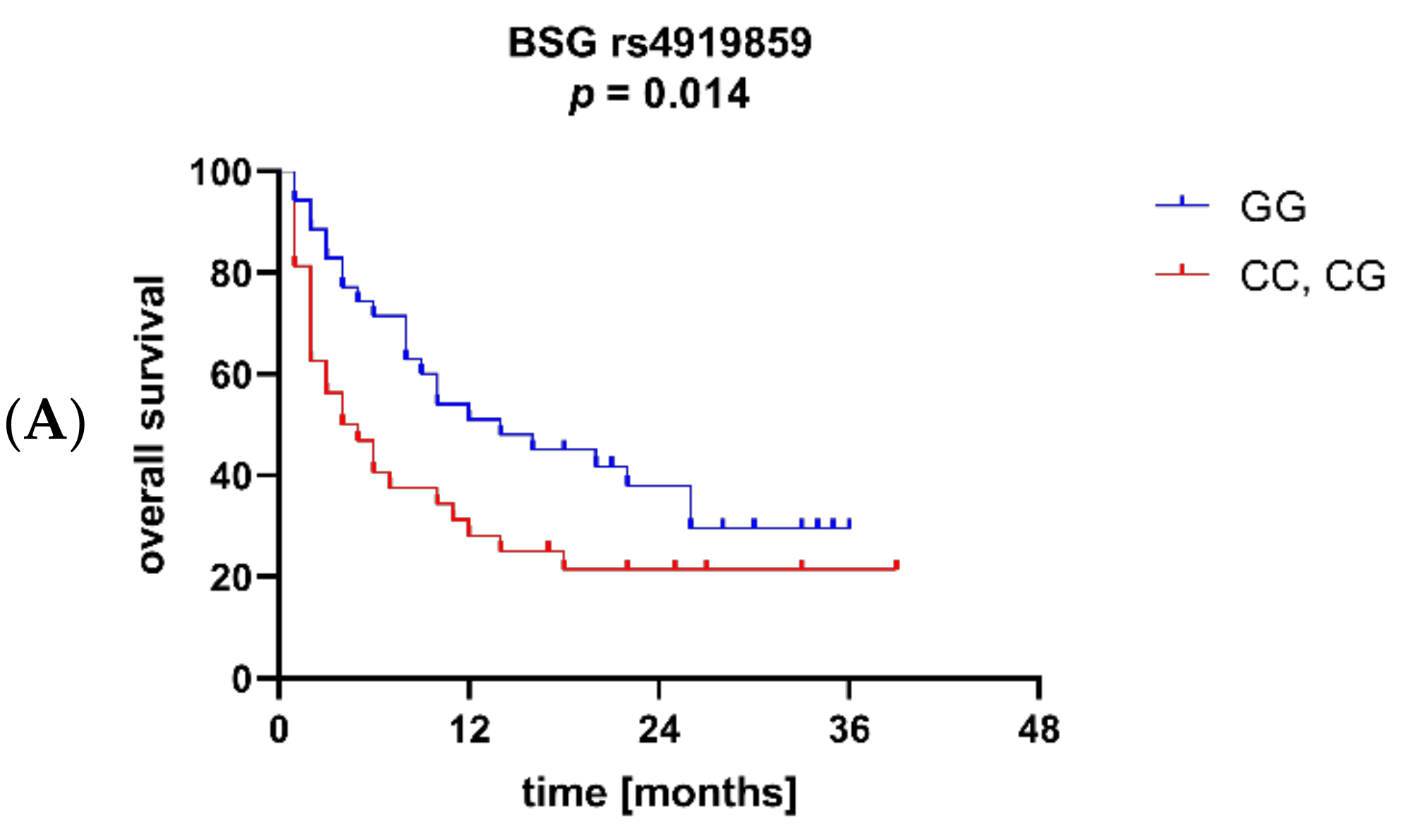
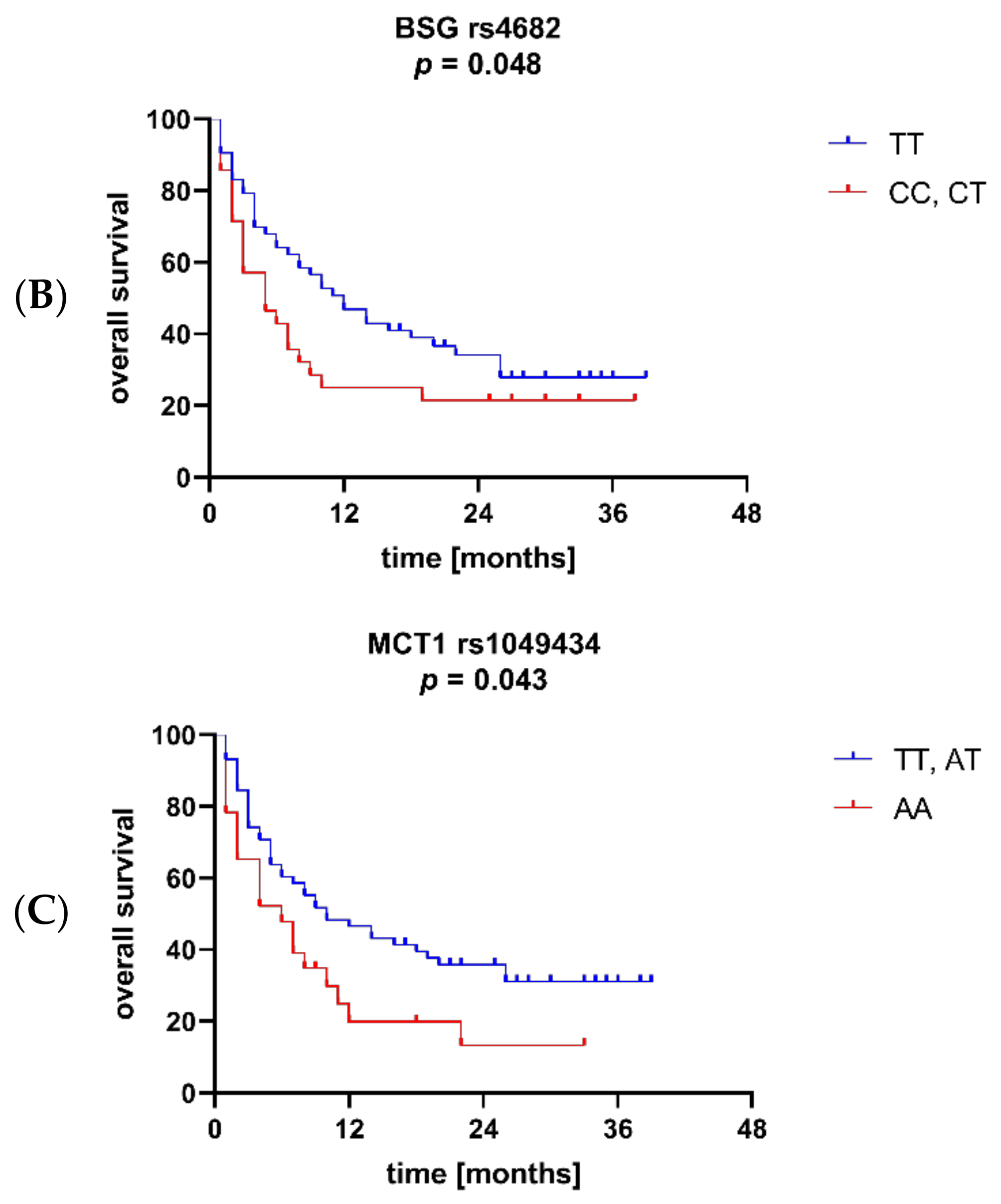
3.3. BSG and MCT1 Genetic Variants Are Associated with Survival and Other Clinical Parameters of AML
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uddin, R.; Darwish, N.; Mousa, S.A. Acute Myeloid Leukemia Mutations and Future Mechanistic Target to Overcome Resistance. Curr. Treat. Options Oncol. 2021, 22, 76. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Doucette, K.; Norsworthy, K. Recent drug approvals for acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.L.; Hills, R.K.; Grimwade, D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2011, 25, 39–51. [Google Scholar] [CrossRef]
- Grass, G.D.; Toole, B.P. How, with whom and when: An overview of CD147-mediated regulatory networks influencing matrix metalloproteinase activity. Biosci. Rep. 2015, 36, e00283. [Google Scholar] [CrossRef] [Green Version]
- Landras, A.; Reger de Moura, C.; Jouenne, F.; Lebbe, C.; Menashi, S.; Mourah, S. CD147 Is a Promising Target of Tumor Progression and a Prognostic Biomarker. Cancers 2019, 11, 1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Xu, C.; Wang, A.; Geng, K.; Honnen, W.; Wang, X.; Bruiners, N.; Singh, S.; Ferrara, F.; D’Angelo, S.; Bradbury, A.; et al. Human Immunodeficiency Viruses Pseudotyped with SARS-CoV-2 Spike Proteins Infect a Broad Spectrum of Human Cell Lines through Multiple Entry Mechanisms. Viruses 2021, 13, 953. [Google Scholar] [CrossRef]
- Riethdorf, S.; Reimers, N.; Assmann, V.; Kornfeld, J.W.; Terracciano, L.; Sauter, G.; Pantel, K. High incidence of EMMPRIN expression in human tumors. Int. J. Cancer 2006, 119, 1800–1810. [Google Scholar] [CrossRef]
- Nabeshima, K.; Iwasaki, H.; Koga, K.; Hojo, H.; Suzumiya, J.; Kikuchi, M. Emmprin (basigin/CD147): Matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol. Int. 2006, 56, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, J.; Chen, L.; Zhong, W.D.; Zhang, Z.; Mi, L.; Zhang, Y.; Liao, C.G.; Bian, H.J.; Jiang, J.L.; et al. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology 2009, 54, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Knutti, N.; Huber, O.; Friedrich, K. CD147 (EMMPRIN) controls malignant properties of breast cancer cells by interdependent signaling of Wnt and JAK/STAT pathways. Mol. Cell. Biochem. 2019, 451, 197–209. [Google Scholar] [CrossRef]
- Marchiq, I.; Albrengues, J.; Granja, S.; Gaggioli, C.; Pouysségur, J.; Simon, M.P. Knock out of the BASIGIN/CD147 chaperone of lactate/H+ symporters disproves its pro-tumour action via extracellular matrix metalloproteases (MMPs) induction. Oncotarget 2015, 6, 24636–24648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halestrap, A.P.; Meredith, D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.M.; Dewhirst, M.W. Tumor metabolism of lactate: The influence and therapeutic potential for MCT and CD147 regulation. Future Oncol. 2010, 6, 127–148. [Google Scholar] [CrossRef] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, D.K.; Arendt, B.K.; Jelinek, D.F. CD147 regulates the expression of MCT1 and lactate export in multiple myeloma cells. Cell Cycle 2013, 12, 3175–3183. [Google Scholar] [CrossRef] [Green Version]
- Granja, S.; Marchiq, I.; Le Floch, R.; Moura, C.S.; Baltazar, F.; Pouysségur, J. Disruption of BASIGIN decreases lactic acid export and sensitizes non-small cell lung cancer to biguanides independently of the LKB1 status. Oncotarget 2015, 6, 6708–6721. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Chang, S.; Jiang, X.; Su, J.; Dong, C.; Liu, X.; Yuan, Z.; Zhang, Z.; Liao, H. RNA interference targeting CD147 inhibits the proliferation, invasiveness, and metastatic activity of thyroid carcinoma cells by down-regulating glycolysis. Int. J. Clin. Exp. Pathol. 2015, 8, 309–318. [Google Scholar] [PubMed]
- Tang, Y.; Nakada, M.T.; Kesavan, P.; McCabe, F.; Millar, H.; Rafferty, P.; Bugelski, P.; Yan, L. Extracellular matrix metalloproteinase inducer stimulates tumor angiogenesis by elevating vascular endothelial cell growth factor and matrix metalloproteinases. Cancer Res. 2005, 65, 3193–3199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.C.; Takahashi, H.; Murai, Y.; Cui, Z.G.; Nomoto, K.; Miwa, S.; Tsuneyama, K.; Takano, Y. Upregulated EMMPRIN/CD147 might contribute to growth and angiogenesis of gastric carcinoma: A good marker for local invasion and prognosis. Br. J. Cancer 2006, 95, 1371–1378. [Google Scholar] [CrossRef]
- Stenzinger, A.; Wittschieber, D.; Von Winterfeld, M.; Goeppert, B.; Kamphues, C.; Weichert, W.; Dietel, M.; Rabien, A.; Klauschen, F. High extracellular matrix metalloproteinase inducer/CD147 expression is strongly and independently associated with poor prognosis in colorectal cancer. Hum. Pathol. 2012, 43, 1471–1481. [Google Scholar] [CrossRef]
- Caudron, A.; Battistella, M.; Feugeas, J.P.; Pages, C.; Basset-Seguin, N.; Mazouz Dorval, S.; Funck Brentano, E.; Sadoux, A.; Podgorniak, M.P.; Menashi, S.; et al. EMMPRIN/CD147 is an independent prognostic biomarker in cutaneous melanoma. Exp. Dermatol. 2016, 25, 618–622. [Google Scholar] [CrossRef]
- Li, H.; Jiang, C.; Wu, D.; Shi, S.; Liao, M.; Wang, J.; Li, Y.; Xu, Z. The prognostic and clinicopathologic characteristics of CD147 and esophagus cancer: A meta-analysis. PLoS ONE 2017, 12, e0180271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arendt, B.K.; Walters, D.K.; Wu, X.; Tschumper, R.C.; Huddleston, P.M.; Henderson, K.J.; Dispenzieri, A.; Jelinek, D.F. Increased expression of extracellular matrix metalloproteinase inducer (CD147) in multiple myeloma: Role in regulation of myeloma cell proliferation. Leukemia 2012, 26, 2286–2296. [Google Scholar] [CrossRef] [Green Version]
- Łacina, P.; Butrym, A.; Mazur, G.; Bogunia-Kubik, K. BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients. Genes 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Fu, J.; Chen, X.; Zhang, Y.; Gu, H.; Bai, Y. CD147 and VEGF co-expression predicts prognosis in patients with acute myeloid leukemia. Jpn. J. Clin. Oncol. 2010, 40, 1046–1052. [Google Scholar] [CrossRef] [Green Version]
- Spinello, I.; Saulle, E.; Quaranta, M.T.; Pasquini, L.; Pelosi, E.; Castelli, G.; Ottone, T.; Voso, M.T.; Testa, U.; Labbaye, C. The small-molecule compound AC-73 targeting CD147 inhibits leukemic cell proliferation, induces autophagy and increases the chemotherapeutic sensitivity of acute myeloid leukemia cells. Haematologica 2019, 104, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Koshikawa, N.; Tomari, T.; Nabeshima, K.; Isobe, T.; Seiki, M. Membrane type 1 matrix metalloproteinase (MT1-MMP/MMP-14) cleaves and releases a 22-kDa extracellular matrix metalloproteinase inducer (EMMPRIN) fragment from tumor cells. J. Biol. Chem. 2006, 281, 37576–37585. [Google Scholar] [CrossRef] [Green Version]
- Albrechtsen, R.; Wewer Albrechtsen, N.J.; Gnosa, S.; Schwarz, J.; Dyrskjøt, L.; Kveiborg, M. Identification of ADAM12 as a Novel Basigin Sheddase. Int. J. Mol. Sci. 2019, 20, 1957. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, S.S.; Mengistab, A.T.; Tauscher, A.N.; LaVail, J.; Basbaum, C. The microvesicle as a vehicle for EMMPRIN in tumor-stromal interactions. Oncogene 2004, 23, 956–963. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Rode, A.; Nicoll, A.; Maczurek, A.E.; Lim, L.; Lim, S.; Angus, P.; Kronborg, I.; Arachchi, N.; Gorelik, A.; et al. Circulating CD147 predicts mortality in advanced hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2016, 31, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.H.; Liu, Y.J.; Tang, L.L.; Wang, S.M.; Yan, G.J.; Liao, L.Q. Plasma soluble cluster of differentiation 147 levels are increased in breast cancer patients and associated with lymph node metastasis and chemoresistance. Hong Kong Med. J. 2018, 24, 252–260. [Google Scholar] [CrossRef] [Green Version]
- Rurali, E.; Perrucci, G.L.; Gaetano, R.; Pini, A.; Moschetta, D.; Gentilini, D.; Nigro, P.; Pompilio, G. Soluble EMMPRIN levels discriminate aortic ectasia in Marfan syndrome patients. Theranostics 2019, 9, 2224–2234. [Google Scholar] [CrossRef] [PubMed]
- Knutti, N.; Kuepper, M.; Friedrich, K. Soluble extracellular matrix metalloproteinase inducer (EMMPRIN, EMN) regulates cancer-related cellular functions by homotypic interactions with surface CD147. FEBS J. 2015, 282, 4187–4200. [Google Scholar] [CrossRef] [Green Version]
- Dratwa, M.; Wysoczanska, B.; Turlej, E.; Anisiewicz, A.; Maciejewska, M.; Wietrzyk, J.; Bogunia-Kubik, K. Heterogeneity of telomerase reverse transcriptase mutation and expression, telomerase activity and telomere length across human cancer cell lines cultured in vitro. Exp. Cell Res. 2020, 396, 112298. [Google Scholar] [CrossRef]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef] [Green Version]
- Beesley, A.H.; Cummings, A.J.; Freitas, J.R.; Hoffmann, K.; Firth, M.J.; Ford, J.; de Klerk, N.H.; Kees, U.R. The gene expression signature of relapse in paediatric acute lymphoblastic leukaemia: Implications for mechanisms of therapy failure. Br. J. Haematol. 2005, 131, 447–456. [Google Scholar] [CrossRef]
- Beesley, A.H.; Weller, R.E.; Kees, U.R. The role of BSG (CD147) in acute lymphoblastic leukaemia and relapse. Br. J. Haematol. 2008, 142, 1000–1002. [Google Scholar] [CrossRef] [PubMed]
- Luczynski, W.; Kowalczuk, O.; Stasiak-Barmuta, A.; Ilendo, E.; Krawczuk-Rybak, M.; Chyczewski, L. Acute lymphoblastic leukemia-derived dendritic cells express tumor associated antigens: PNPT1, PMPCB, RHAMM, BSG and ERCC1. Neoplasma 2009, 56, 428–434. [Google Scholar] [CrossRef]
- De Vries, J.F.; Te Marvelde, J.G.; Wind, H.K.; Van Dongen, J.J.; Van der Velden, V.H. The potential use of basigin (CD147) as a prognostic marker in B-cell precursor acute lymphoblastic leukaemia. Br. J. Haematol. 2010, 150, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Saulle, E.; Spinello, I.; Quaranta, M.T.; Pasquini, L.; Pelosi, E.; Iorio, E.; Castelli, G.; Chirico, M.; Pisanu, M.E.; Ottone, T.; et al. Targeting Lactate Metabolism by Inhibiting MCT1 or MCT4 Impairs Leukemic Cell Proliferation, Induces Two Different Related Death-Pathways and Increases Chemotherapeutic Sensitivity of Acute Myeloid Leukemia Cells. Front. Oncol. 2021, 10, 621458. [Google Scholar] [CrossRef]
- Bolomsky, A.; Hübl, W.; Spada, S.; Müldür, E.; Schlangen, K.; Heintel, D.; Rocci, A.; Weißmann, A.; Fritz, V.; Willheim, M.; et al. IKAROS expression in distinct bone marrow cell populations as a candidate biomarker for outcome with lenalidomide-dexamethasone therapy in multiple myeloma. Am. J. Hematol. 2017, 92, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi Najafabadi, M.; Shamsasenjan, K.; Akbarzadehalaleh, P. Angiogenesis Status in Patients with Acute Myeloid Leukemia: From Diagnosis to Post-hematopoietic Stem Cell Transplantation. Int. J. Organ Transplant. Med. 2017, 8, 57–67. [Google Scholar]
- Cui, H.Y.; Guo, T.; Wang, S.J.; Zhao, P.; Dong, Z.S.; Zhang, Y.; Jiang, J.L.; Chen, Z.N.; Yu, X.L. Dimerization is essential for HAb18G/CD147 promoting tumor invasion via MAPK pathway. Biochem. Biophys. Res. Commun. 2012, 419, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.M.; Kuo, P.J.; Lin, C.Y.; Chin, Y.T.; Lin, H.L.; Chiu, H.C.; Fu, M.; Fu, E. CD147 self-regulates matrix metalloproteinase-2 release in gingival fibroblasts after coculturing with U937 monocytic cells. J. Periodontol. 2020, 91, 651–660. [Google Scholar] [CrossRef]
- Butrym, A.; Łacina, P.; Kuliczkowski, K.; Bogunia-Kubik, K.; Mazur, G. Genetic variation of the gene coding for microRNA-204 (miR-204) is a risk factor in acute myeloid leukaemia. BMC Cancer 2018, 18, 107. [Google Scholar] [CrossRef] [Green Version]
- Butrym, A.; Łacina, P.; Bogunia-Kubik, K.; Mazur, G. ABCC3 and GSTM5 gene polymorphisms affect overall survival in Polish acute myeloid leukaemia patients. Curr. Probl. Cancer 2021, 45, 100729. [Google Scholar] [CrossRef]
- Weng, Y.; Chen, T.; Ren, J.; Lu, D.; Liu, X.; Lin, S.; Xu, C.; Lou, J.; Chen, X.; Tang, L. The Association Between Extracellular Matrix Metalloproteinase Inducer Polymorphisms and Coronary Heart Disease: A Potential Way to Predict Disease. DNA Cell Biol. 2020, 39, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Futagi, Y.; Kobayashi, M.; Ogura, J.; Iseki, K. Functional characterization of 5-oxoproline transport via SLC16A1/MCT1. J. Biol. Chem. 2015, 290, 2303–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilherme, J.; Bosnyák, E.; Semenova, E.A.; Szmodis, M.; Griff, A.; Móra, Á.; Almási, G.; Trájer, E.; Udvardy, A.; Kostryukova, E.S.; et al. The MCT1 gene Glu490Asp polymorphism (rs1049434) is associated with endurance athlete status, lower blood lactate accumulation and higher maximum oxygen uptake. Biol. Sport 2021, 38, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Massidda, M.; Flore, L.; Kikuchi, N.; Scorcu, M.; Piras, F.; Cugia, P.; Cięszczyk, P.; Tocco, F.; Calò, C.M. Influence of the MCT1-T1470A polymorphism (rs1049434) on repeated sprint ability and blood lactate accumulation in elite football players: A pilot study. Eur. J. Appl. Physiol. 2021, 121, 3399–3408. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Guo, X.; Chen, Y.; Liu, X.; Tu, J.; Xing, J.; Chen, Z.; Ji, J.; He, X. Polymorphisms of monocarboxylate transporter genes are associated with clinical outcomes in patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Labbaye, C.; Castelli, G.; Pelosi, E. Oxidative stress and hypoxia in normal and leukemic stem cells. Exp. Hematol. 2016, 44, 540–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Data | Serum Study Group Range and Median (n = 37) 1 | Genetic Study Group Range and Median (n = 92) 1 |
|---|---|---|
| age | 25–93, med = 62 | 20–93, med = 61 |
| bone marrow blasts (%) | 4–95, med = 68 | 20–98, med = 65 |
| white blood cell count (G/L) | 0.8–510.5, med = 22.7 | 0.7–510.5, med = 21 |
| haemoglobin (g/dL) | 6.0–11.4, med = 8.8 | 6.1–19.5, med = 9.2 |
| platelets (G/L) | 5–393, med = 44.5 | 2–460, med = 52 |
| lactate dehydrogenase (U/L) | 168–2170, med = 446.5 | 26–2535, med = 369 |
| C-reactive protein (mg/L) | 2.3–115.4, med = 36.3 | 1.1–359, med = 25 |
| AML Patients | Healthy Individuals | |
|---|---|---|
| BSG rs4919859 | ||
| CC | 18 (19.6%) | 19 (14.1%) |
| CG | 35 (38.0%) | 54 (40.0%) |
| GG | 39 (43.4%) | 62 (45.9%) |
| BSG rs4682 | ||
| CC | 7 (7.6%) | 4 (3.0%) |
| CT | 26 (28.3%) | 35 (25.9%) |
| TT | 59 (64.1%) | 96 (71.1%) |
| BSG rs8637 | ||
| AA | 30 (32.6%) | 47 (34.8%) |
| AG | 45 (48.9%) | 61 (45.2%) |
| GG | 17 (18.5%) | 27 (20.0%) |
| BSG rs8259 | ||
| TT | 50 (54.3%) | 78 (57.8%) |
| TA | 31 (33.7%) | 49 (36.3%) |
| AA | 11 (12.0%) | 8 (5.9%) |
| MCT1 rs1049434 | ||
| AA | 28 (30.4%) | 54 (40.0%) |
| AT | 45 (48.9%) | 58 (43.0%) |
| TT | 19 (20.7%) | 23 (17.0%) |
| MCT1 rs9429505 | ||
| AA | 52 (56.5%) | 67 (49.6%) |
| AG | 34 (37.0%) | 60 (44.4%) |
| GG | 6 (6.5%) | 8 (5.9%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łacina, P.; Butrym, A.; Turlej, E.; Stachowicz-Suhs, M.; Wietrzyk, J.; Mazur, G.; Bogunia-Kubik, K. BSG (CD147) Serum Level and Genetic Variants Are Associated with Overall Survival in Acute Myeloid Leukaemia. J. Clin. Med. 2022, 11, 332. https://doi.org/10.3390/jcm11020332
Łacina P, Butrym A, Turlej E, Stachowicz-Suhs M, Wietrzyk J, Mazur G, Bogunia-Kubik K. BSG (CD147) Serum Level and Genetic Variants Are Associated with Overall Survival in Acute Myeloid Leukaemia. Journal of Clinical Medicine. 2022; 11(2):332. https://doi.org/10.3390/jcm11020332
Chicago/Turabian StyleŁacina, Piotr, Aleksandra Butrym, Eliza Turlej, Martyna Stachowicz-Suhs, Joanna Wietrzyk, Grzegorz Mazur, and Katarzyna Bogunia-Kubik. 2022. "BSG (CD147) Serum Level and Genetic Variants Are Associated with Overall Survival in Acute Myeloid Leukaemia" Journal of Clinical Medicine 11, no. 2: 332. https://doi.org/10.3390/jcm11020332






