Comparing a Fully Automated Cephalometric Tracing Method to a Manual Tracing Method for Orthodontic Diagnosis
Abstract
:1. Introduction
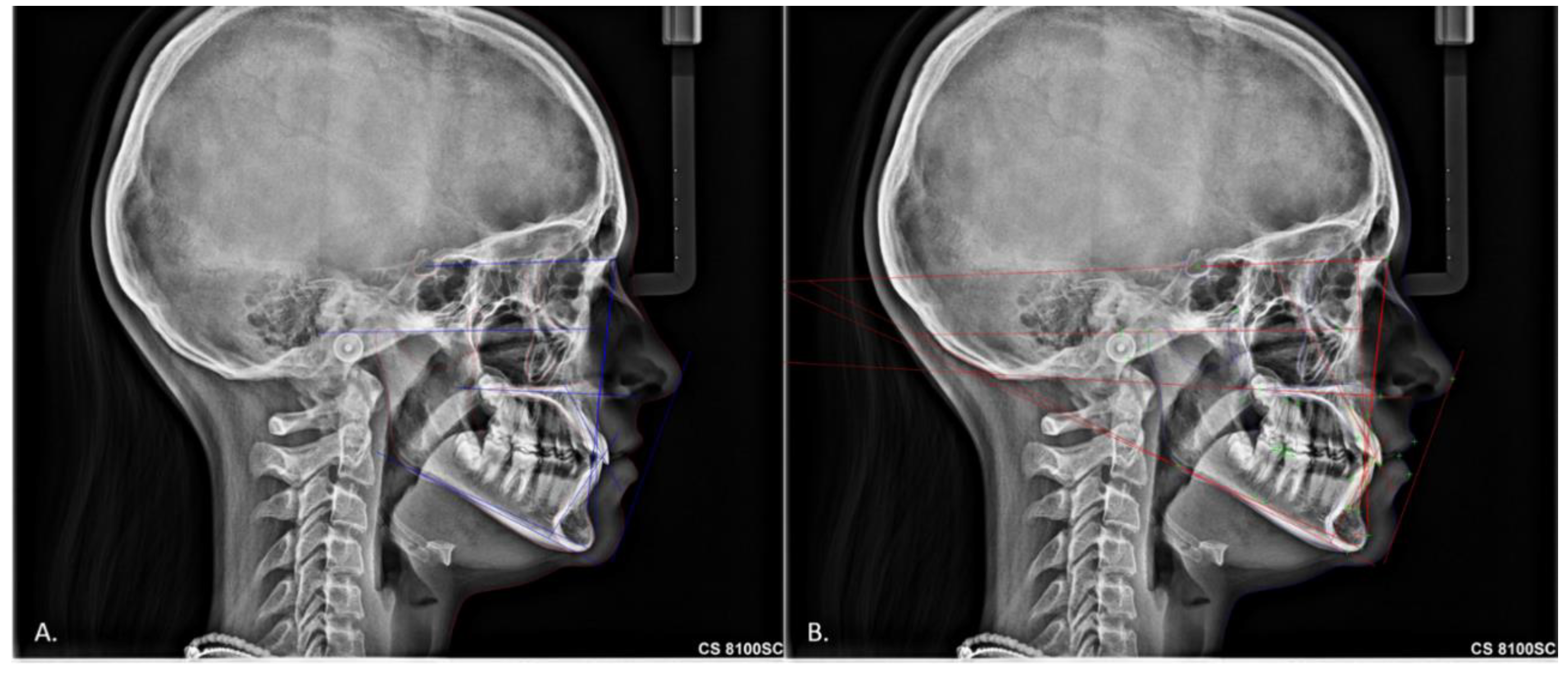
2. Materials and Methods
Statistical Analysis
3. Results
3.1. American Board of Orthodontics Cephalometric Analysis
3.2. European Board of Orthodontics Cephalometric Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Broadbent, B.H. A new x-ray technique and its application to orthodontia. Angle Orthod. 1931, 1, 45–66. [Google Scholar]
- Naoumova, J.; Lindman, R. A comparison of manual traced images and corresponding scanned radiographs digitally traced. Eur. J. Orthod. 2009, 31, 247–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thurzo, A.; Urbanová, W.; Novák, B.; Czako, L.; Siebert, T.; Stano, P.; Mareková, S.; Fountoulaki, G.; Kosnáčová, H.; Varga, I. Where Is the Artificial Intelligence Applied in Dentistry? Systematic Review and Literature Analysis. Healthcare 2022, 10, 1269. [Google Scholar] [CrossRef] [PubMed]
- Tsolakis, I.A.; Gizani, S.; Panayi, N.; Antonopoulos, G.; Tsolakis, A.I. Three-Dimensional Printing Technology in Orthodontics for Dental Models: A Systematic Review. Children 2022, 9, 1106. [Google Scholar] [CrossRef]
- Tsolakis, I.A.; Gizani, S.; Tsolakis, A.I.; Panayi, N. Three-Dimensional-Printed Customized Orthodontic and Pedodontic Appliances: A Critical Review of a New Era for Treatment. Children 2022, 9, 1107. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, H.M.; Nadimi, M.; Rohban, M.H.; Shamsoddin, E.; Lee, V.Y.; Motamediane, S.R. Machine learning and orthodontics, current trends and the future opportunities: A scoping review. Am. J. Orthod. Dentofac. Orthop. 2021, 160, 170–192. [Google Scholar] [CrossRef]
- Arik, S.; Ibragimov, B.; Xing, L. Fully automated quantitative cephalometry using convolutional neural networks. J. Med. Imaging 2017, 4, 014501. [Google Scholar] [CrossRef]
- Park, J.-H.; Hwang, H.-W.; Moon, J.-H.; Yu, Y.; Kim, H.; Her, S.-B.; Srinivasan, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.-J. Automated identification of cephalometric landmarks: Part 1—Comparisons between the latest deep-learning methods YOLOV3 and SSD. Angle Orthod. 2019, 89, 903–909. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-H.; Vandaele, R.; Mirzaalian, H.; Chen, C.; Li, W.-C.; Zheng, G.; Jodogne, S.; Chang, S.-W.; Wang, C.W.; Maree, R.; et al. Evaluation and Comparison of Anatomical Landmark Detection Methods for Cephalometric X-Ray Images: A Grand Challenge. IEEE Trans. Med. Imaging 2015, 34, 1590–1900. [Google Scholar] [CrossRef]
- Lindner, C.; Wang, C.-W.; Huang, C.-T.; Li, C.-H.; Chang, S.-W.; Cootes, T.F. Fully Automatic System for Accurate Localisation and Analysis of Cephalometric Landmarks in Lateral Cephalograms. Sci. Rep. 2016, 6, 33581. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Li, J.; Zhang, Y.; Zou, B. Automatic Analysis of Lateral Cephalograms Based on Multiresolution Decision Tree Regression Voting. J. Health Eng. 2018, 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-S.; Adhikari, S.; Liu, L.; Jeong, H.-G.; Kim, H.; Yoon, S.-J. Osteoporosis detection in panoramic radiographs using a deep convolutional neural network-based computer-assisted diagnosis system: A preliminary study. Dentomaxillofacial Radiol. 2019, 48, 20170344. [Google Scholar] [CrossRef]
- Ekert, T.; Krois, J.; Meinhold, L.; Elhennawy, K.; Emara, R.; Golla, T.; Schwendicke, F. Deep Learning for the Radiographic Detection of Apical Lesions. J. Endod. 2019, 45, 917–922.e5. [Google Scholar] [CrossRef]
- Moutselos, K.; Berdouses, E.; Oulis, C.; Maglogiannis, I. Recognizing Occlusal Caries in Dental Intraoral Images Using Deep Learning. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 1617–1620. [Google Scholar] [CrossRef]
- Aubreville, M.; Knipfer, C.; Oetter, N.; Jaremenko, C.; Rodner, E.; Denzler, J.; Bohr, C.; Neumann, H.; Stelzle, F.; Maier, A. Automatic Classification of Cancerous Tissue in Laserendomicroscopy Images of the Oral Cavity using Deep Learning. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Yan, H.; Xia, J.; Zhu, L.; Zhang, T.; Zhu, Z.; Lou, X.; Sun, G.; Dong, M. Deep convolutional neural networks for tongue squamous cell carcinoma classification using Raman spectroscopy. Photodiagnosis Photodyn. Ther. 2019, 26, 430–435. [Google Scholar] [CrossRef]
- Torosdagli, N.; Liberton, D.K.; Verma, P.; Sincan, M.; Lee, J.S.; Bagci, U. Deep Geodesic Learning for Segmentation and Anatomical Landmarking. IEEE Trans. Med. Imaging 2019, 38, 919–931. [Google Scholar] [CrossRef]
- Egger, J.; Pfarrkirchner, B.; Gsaxner, C.; Lindner, L.; Schmalstieg, D.; Wallner, J. Fully Convolutional Mandible Segmentation on a valid Ground- Truth Dataset. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 656–660. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, L.; Xia, W.; Zhang, B.; Qiu, B.; Gao, X. Multiple supervised residual network for osteosarcoma segmentation in CT images. Comput. Med. Imaging Graph. 2018, 63, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Poedjiastoeti, W.; Suebnukarn, S. Application of Convolutional Neural Network in the Diagnosis of Jaw Tumors. Health Informatics Res. 2018, 24, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Casalegno, F.; Newton, T.; Daher, R.; Abdelaziz, M.; Lodi-Rizzini, A.; Schürmann, F.; Krejci, I.; Markram, H. Caries detection with near-infrared transillumination using deep learning. J. Dent. Res. 2019, 98, 1227–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuzoff, D.V.; Tuzova, L.N.; Bornstein, M.M.; Krasnov, A.S.; Kharchenko, M.A.; Nikolenko, S.I.; Sveshnikov, M.M.; Bednenko, G.B. Tooth detection and numbering in panoramic radiographs using convolutional neural networks. Dentomaxillofac. Radiol. 2019, 48, 20180051. [Google Scholar] [CrossRef]
- Hwang, H.W.; Park, J.H.; Moon, J.H.; Yu, Y.; Kim, H.; Her, S.B.; Srinivasan, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.J. Automated identification of cephalometric landmarks: Part 2-Might it be better than human? Angle Orthod. 2020, 90, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-K.; Chen, Y.-J.; Yao, C.-C.J.; Chang, H.-F. Enhanced speed and precision of measurement in a computer-assisted digital cephalometric analysis system. Angle Orthod. 2004, 74, 501–507. [Google Scholar] [CrossRef]
- Erkan, M.; Gurel, H.G.; Nur, M.; Demirel, B. Reliability of four different computerized cephalometric analysis programs. Eur. J. Orthod. 2012, 34, 318–321. [Google Scholar] [CrossRef] [Green Version]
- Uysal, T.; Baysal, A.; Yagci, A. Evaluation of speed, repeatability, and reproducibility of digital radiography with manual versus computer-assisted cephalometric analyses. Eur. J. Orthod. 2009, 31, 523–528. [Google Scholar] [CrossRef]
- Meriç, P.; Naoumova, J. Web-based Fully Automated Cephalometric Analysis: Comparisons between App-aided, Computerized, and Manual Tracings. Turk. J. Orthod. 2020, 33, 142–149. [Google Scholar] [CrossRef]
- Bulatova, G.; Kusnoto, B.; Grace, V.; Tsay, T.P.; Avenetti, D.M.; Sanchez, F.J.C. Assessment of automatic cephalometric landmark identification using artificial intelligence. Orthod. Craniofac. Res. 2021, 24, 37–42. [Google Scholar] [CrossRef] [PubMed]
| Parameter | ICC (95% CI) | |
|---|---|---|
| Automatic | Manual | |
| ANB | 1.00 (1.00, 1.00) | 0.99 (0.97, 0.99) |
| ANPg | 1.00 (1.00, 1.00) | 0.99 (0.97, 0.99) |
| ANS-PNS/GoGn | 1.00 (1.00, 1.00) | 0.97 (0.93, 0.99) |
| FMA (MP-FH) | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) |
| IMPA (L1-MP) | 1.00 (1.00, 1.00) | 0.98 (0.94, 0.99) |
| L1-NB | 1.00 (1.00, 1.00) | 0.97 (0.92, 0.99) |
| L1/APg | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) |
| L1/GoGn | 1.00 (1.00, 1.00) | 0.98 (0.95, 0.99) |
| SN-GoGn | 1.00 (1.00, 1.00) | 0.98 (0.95, 0.99) |
| SN/ANS-PNS | 1.00 (1.00, 1.00) | 0.96 (0.89, 0.98) |
| SNA | 1.00 (1.00, 1.00) | 0.95 (0.88, 0.98) |
| SNB | 1.00 (1.00, 1.00) | 0.97 (0.92, 0.99) |
| SNMP | 1.00 (1.00, 1.00) | 0.98 (0.94, 0.99) |
| SNPg | 1.00 (1.00, 1.00) | 0.98 (0.96, 0.99) |
| U1-NA | 1.00 (1.00, 1.00) | 0.99 (0.99, 1.00) |
| U1-L1 | 1.00 (1.00, 1.00) | 0.99 (0.99, 1.00) |
| U1-SN | 1.00 (1.00, 1.00) | 0.99 (0.99, 1.00) |
| U1/ANS-PNS | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) |
| Parameter | ICC (95% CI) |
|---|---|
| ANB | 0.85 (0.70, 0.97) |
| ANPg | 0.77 (0.61, 0.91) |
| ANS-PNS/GoGn | 0.72 (0.45, 0.91) |
| FMA (MP-FH) | 0.73 (0.47, 0.85) |
| IMPA (L1-MP) | 0.70 (0.49, 0.87) |
| L1-NB | 0.74 (0.76, 0.93) |
| L1/APg | 0.75 (0.57, 0.89) |
| L1/GoGn | 0.78 (0.60, 0.93) |
| SN-GoGn | 0.89 (0.72, 0.92) |
| SN/ANS-PNS | 0.77 (0.59, 0.93) |
| SNA | 0.74 (0.55, 0.90) |
| SNB | 0.78 (0.60, 0.94) |
| SNMP | 0.89 (0.73, 0.93) |
| SNPg | 0.92 (0.88, 0.94) |
| U1-NA | 0.79 (0.70, 0.86) |
| U1-L1 | 0.70 (0.54, 0.81) |
| U1-SN | 0.76 (0.61, 0.88) |
| U1/ANS-PNS | 0.72 (0.53, 0.89) |
| Automatic | Manual | Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Mean (SD) | min | max | Mean (SD) | min | max | Auto-Manual | p-Value * |
| ANB | 3.8 (2.7) | 0.0 | 14.0 | 3.6 (2.4) | 0.1 | 9.5 | 0.2 | 0.517 |
| ANPg | 5.8 (5.0) | 0.0 | 29.0 | 5.4 (3.8) | 0.1 | 16.5 | 0.4 | 0.467 |
| ANS-PNS/GoGn | 27.1 (4.3) | 17.7 | 45.0 | 24.1 (5.2) | 9.4 | 33.8 | 3 | <0.001 * |
| FMA (MP-FH) | 30.2 (5.6) | 15.9 | 49.0 | 28.1 (5.3) | 14.9 | 39.4 | 2.1 | <0.001 * |
| IMPA (L1-MP) | 87.1 (7.6) | 61.4 | 104.8 | 91.1 (7.8) | 71.3 | 110.1 | −4 | <0.001 * |
| L1-NB | 156.8 (16.1) | 13.7 | 174.8 | 156.3 (6.6) | 139.4 | 176.4 | 0.5 | 0.150 |
| L1/APg | 155.3 (27.0) | 4.1 | 173.9 | 156.6 (15.4) | 13.2 | 173.2 | −1.3 | 0.945 |
| L1/GoGn | 90.6 (7.5) | 64.1 | 107.0 | 93.2 (8.0) | 74.2 | 112.1 | −2.6 | 0.003 |
| SN-GoGn | 33.1 (6.6) | 19.0 | 55.4 | 31.7 (5.4) | 16.3 | 42.4 | 1.4 | 0.021 |
| SN/ANS-PNS | 7.0 (4.8) | 0.0 | 18.8 | 7.7 (3.4) | 1.1 | 17.0 | −0.7 | 0.125 |
| SNA | 82.4 (6.5) | 67.1 | 106.2 | 81.7 (3.4) | 73.4 | 91.5 | 0.7 | 0.246 |
| SNB | 78.9 (6.5) | 63.2 | 98.8 | 78.2 (3.8) | 71.4 | 90.0 | 0.7 | 0.256 |
| SNMP | 35.6 (6.6) | 21.7 | 58.5 | 34.4 (5.5) | 19.5 | 45.4 | 1.2 | 0.061 |
| SNPg | 98.5 (7.8) | 82.4 | 120.1 | 98.2 (7.9) | 80.5 | 119.5 | 0.3 | 0.252 |
| U1-NA | 35.9 (5.5) | 30.4 | 41.4 | 35.4 (3.4) | 32 | 38.8 | 0.5 | 0.014 |
| U1-L1 | 131.9 (11.1) | 107.0 | 167.6 | 128.6 (10.5) | 101.2 | 157.0 | 3.3 | <0.001 * |
| U1-SN | 105.6 (7.7) | 88.2 | 122.6 | 105.9 (8.0) | 90.2 | 124.6 | −0.3 | 0.629 |
| U1/ANS-PNS | 111.6 (5.5) | 96.4 | 125.8 | 112.8 (9.2) | 54.2 | 130.9 | −1.2 | 0.195 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsolakis, I.A.; Tsolakis, A.I.; Elshebiny, T.; Matthaios, S.; Palomo, J.M. Comparing a Fully Automated Cephalometric Tracing Method to a Manual Tracing Method for Orthodontic Diagnosis. J. Clin. Med. 2022, 11, 6854. https://doi.org/10.3390/jcm11226854
Tsolakis IA, Tsolakis AI, Elshebiny T, Matthaios S, Palomo JM. Comparing a Fully Automated Cephalometric Tracing Method to a Manual Tracing Method for Orthodontic Diagnosis. Journal of Clinical Medicine. 2022; 11(22):6854. https://doi.org/10.3390/jcm11226854
Chicago/Turabian StyleTsolakis, Ioannis A., Apostolos I. Tsolakis, Tarek Elshebiny, Stefanos Matthaios, and J. Martin Palomo. 2022. "Comparing a Fully Automated Cephalometric Tracing Method to a Manual Tracing Method for Orthodontic Diagnosis" Journal of Clinical Medicine 11, no. 22: 6854. https://doi.org/10.3390/jcm11226854







